Constraint File
Metadata
This document was edited by Colin Smith on 12/4/2008. Yi Liu created the initial page. Thanks Oliver and Firas for providing information.
File Formats
Constraints can be specified using two different file formats, line-based and section-based. The line-based format is:
Constraint_Type1 Constraint_Def1
Constraint_Type2 Constraint_Def2
...
Generally speaking, the Constraint_Type will contain a type, defining what sort of value to be constrained (distance, angle, dihedral, etc), and a series of atom and/or residue labels defining a specific quality to be constrained. Residue numbers are assumed to be in Rosetta numbering (from 1, no gaps), not PDB numbering. If you want PDB numbering, pass the chain letter immediately after the residue number (no spaces), residue 30 of chain A would be "30A", even if it is the first residue in the PDB (Rosetta numbering "1").
The Constraint_Def will define function by which the constraint is constrained, to answer the question: what should the score of the constraint be when the constrained value has a deviation of X units?
Constraint Types
Constraint types are all implemented as subclasses of the core::scoring::constraints::Constraint class.
Single constraints
Single constraints restrain the value of a single metric
-
AtomPair:
AtomPair Atom1_Name Atom1_ResNum Atom2_Name Atom2_ResNum Func_Type Func_Def- Constrains a distance between Atom1 and Atom2. AtomPairConstraint is compatible with PDB numbering.
-
Angle:
Angle Atom1_Name Atom1_ResNum Atom2_Name Atom2_ResNum Atom3_Name Atom3_ResNum Func_Type Func_Def- Angle between Atom2->Atom1 vector and Atom2->Atom3 vector; the angle (passed as a value to the Func) appears to be measured in radians
-
Dihedral:
Dihedral Atom1_Name Atom1_ResNum Atom2_Name Atom2_ResNum Atom3_Name Atom3_ResNum Atom4_Name Atom4_ResNum Func_Type Func_Def- Dihedral angle of Atom1->Atom2->Atom3->Atom4. Dihedral is measured in radians on -pi -> pi.
-
CoordinateConstraint:
CoordinateConstraint Atom1_Name Atom1_ResNum Atom2_Name Atom2_ResNum Atom1_target_X_coordinate Atom1_target_Y_coordinate Atom1_target_Z_coordinate Func_Type Func_Def- Constrain Atom1 to the XYZ position listed. Atom2 is used as a reference atom to determine when Atom1 has moved (so that Rosetta knows when to rescore it) - pick at atom that Atom1 will move relative to. CoordinateConstraint is compatible with PDB numbering.
-
LocalCoordinateConstraint:
LocalCoordinateConstraint Atom1_Name Atom1_ResNum Atom2_Name Atom3_Name Atom4_Name Atom234_ResNum Atom1_target_X_coordinate Atom1_target_Y_coordinate Atom1_target_Z_coordinate Func_Type Func_Def- Constrain Atom1 to the XYZ position listed, relative to the coordinate frame defined by atoms 2/3/4 instead of the origin. LocalCoordinateConstraint is compatible with PDB numbering.
-
AmbiguousNMRDistance:
AmbiguousNMRDistance Atom1_Name Atom1_ResNum Atom2_Name Atom2_ResNum Func_Type Func_Def- Distance between Atom1 and Atom2. The difference from AtomPairConstraint is that atom names are specially parsed to detect ambiguous hydrogens, which are either experimentally ambiguous or rotationally identical (like methyl hydrogens). The constraint applies to any hydrogens equivalent to the named hydrogen. The logic for determining which hydrogens are which is in src/core/scoring/constraints/AmbiguousNMRDistanceConstraints.cc:parse_NMR_name.
-
SiteConstraint:
SiteConstraint Atom1_Name Atom1_ResNum Opposing_chain Func_Type Func_Def- Constraint that a residue interacts with some other chain - roughly, that it is (or is not) in a binding site. The atom and resnum identify which atom is being checked for interactions with the opposing chain. Notice that "Constraint" is irregularly in its tag.
BigBin:
BigBin ResNum Bin
Nested constraints
Nested constraints take as their parameters one or more other constraints, and allow optimization across multiple constraints. Typically in constraint files these are listed across multiple lines, with the name of the constraint opening the block of sub-constraints, and a line starting with "END" or "End" ending the block.
-
MultiConstraint:
MultiConstraint Constraint_Type1 Constraint_Def1 [Constraint_Type2 Constraint_Def2 [...]] END
- Sum of all specified constraints
-
AmbiguousConstraint:
AmbiguousConstraint Constraint_Type1 Constraint_Def1 [Constraint_Type2 Constraint_Def2 [...]] END
- Lowest of all specified constraints
-
KofNConstraint:
KofNConstraint k Constraint_Type1 Constraint_Def1 [Constraint_Type2 Constraint_Def2 [...]] END
- Value is the sum of the "k" lowest energy constraints
Function Types
Functions are listed as "Func_Type Func_Def".
-
HARMONIC x0 sd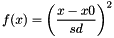
-
CIRCULARHARMONIC x0 sd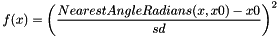
-
BOUNDED lb ub sd rswitch tag- Note: Setting
rswitchto anything other than 0.5 will create a discontinuity in the derivative.rswitchandtagshould not be treated as optional.

- Note: Setting
-
GAUSSIANFUNC mean sd tag- Note:
tagis NOT optional, as for BoundFunc/BOUNDED. If `tag = NOLOG, it triggers some undocumented behavior involving a logarithm of some sort.
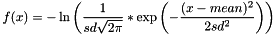
- Note:
-
SOGFUNC n_funcs [mean1 sdev1 weight1 [mean2 sdev2 weight2 [...]]]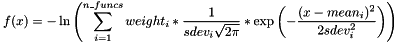
-
MIXTUREFUNC anchor gaussian_param exp_param mixture_param bg_mean bg_sd- parameters: representing the mean of h(r), representing the standard deviation of h(r)
-
CONSTANTFUNC return_val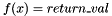
-
SCALARWEIGHTEDFUNC weight Func_Type Func_Def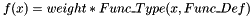
-
SUMFUNC n_funcs Func_Type1 Func_Def1 [Func_Type2 Func_Def2 [...]]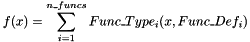
-
SPLINE histogram_file_path experimental_value weight bin_size- Note: This function reads in any histogram and creates a cubic spline over it using the Rosetta SplineGenerator. The full path to the file must be speified. The function assumes that all bin sizes are the same, even though you must specify it for each line in the cst file. If using RosettaEPR knowledge-based potential, replace <filename> with EPR_DISTANCE, and it will read in the EPR distance histogram from the Rosetta database. See example below for using with EPR knowledge-based potential.
-
FADE lb ub d wd [ wo ]- This is meant to basically be a smoothed "square well" bonus of
wdbetween the boundarieslbandub. An optional offsetwo(default 0) can be added to the whole function; this is useful if you want to make the function be zero in the 'golden range' and then give a penalty elsewhere (e.g., specify wd of -20 and wo of +20). To make sure the function and its derivative are continuous, the function is connected by cubic splines in the boundary regions in slivers of width d, betweenlbtolb+dand betweenub-dtoub:
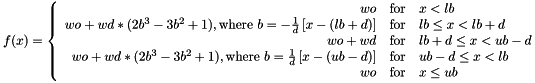
- This is meant to basically be a smoothed "square well" bonus of
-
SIGMOID x0 m- Two arguments; x0 is the center of the sigmoid func and m is its slope. It has hardcoded min/max of 0.5. The functional form is (1/(1+exp(-m*( x-x0 ))) - 0.5.
Function types are all implemented as subclasses of the core::scoring::constraints::Func class.
Sample Files
AtomPair CZ 20 CA 6 GAUSSIANFUNC 5.54 2.0
AtomPair CZ 20 CA 54 GAUSSIANFUNC 5.27 2.0
AtomPair CZ 20 CA 50 GAUSSIANFUNC 5.26 2.0
AtomPair CZ 20 CA 10 GAUSSIANFUNC 4.81 2.0
AtomPair CZ 20 CA 41 GAUSSIANFUNC 9.90 2.0
AtomPair SG 5 V1 32 HARMONIC 0.0 0.2
Angle CB 5 SG 5 ZN 32 HARMONIC 1.95 0.35
AtomPair SG 8 V2 32 HARMONIC 0.0 0.2
Angle CB 8 SG 8 ZN 32 HARMONIC 1.95 0.35
AtomPair NE2 13 V3 32 HARMONIC 0.0 0.2
Angle CD2 13 NE2 13 ZN 32 HARMONIC 2.09 0.35
Dihedral CG 13 CD2 13 NE2 13 ZN 32 CIRCULARHARMONIC 3.14 0.35
AtomPair SG 18 V4 32 HARMONIC 0.0 0.2
Angle CB 18 SG 18 ZN 32 HARMONIC 1.95 0.35
#discourages residue 16, chain B, from interacting with chain A; encourages residue 47
SiteConstraint CA 16B A SIGMOID 5.0 -2.0
SiteConstraint CA 47B A SIGMOID 5.0 2.0
AtomPair H 6 HA 134 BOUNDED 1.80 5.25 0.50 NOE ;dist 5.000 1.800
AtomPair HA 7 HA 132 HARMONIC 1.2 0.2
AtomPair CB 31 CB 43 SPLINE EPR_DISTANCE 6.0 4.0 0.5
AtomPair CB 29 CB 62 SPLINE EPR_DISTANCE 15.0 4.0 0.5
Code for Reading Constraints
There are different sets of functions for reading full-atom and non-full-atom constraints, as indicated by the respective functions listed below. The only current difference between the functions are which command line arguments are read. The values of the arguments are processed identically.
To use constraints, both the scoring function and pose objects should be updated. The functions for adding constraints to the scoring function are:
- core::scoring::constraints::add_fa_constraints_from_cmdline_to_scorefxn()
- core::scoring::constraints::add_constraints_from_cmdline_to_scorefxn()
Currently these functions only set the weights of the atom_pair_constraint, angle_constraint, and dihedral_constraint score function terms to the value of either the -constraints:cst_fa_weight or -constraints:cst_weight command line argument.
The functions for adding constraints to the pose object are:
- core::scoring::constraints::add_fa_constraints_from_cmdline_to_pose()
- core::scoring::constraints::add_constraints_from_cmdline_to_pose()
These functons read a random constraint file from the list defined by either the -constraints:cst_fa_file or -constraints:cst_file argument.
There are also convenience functions for doing both at once:
- core::scoring::constraints::add_fa_constraints_from_cmdline()
- core::scoring::constraints::add_constraints_from_cmdline()
Other Constraint Types
These constraint types cannot currently be specified in a file. They need to have read_def methods implemented and be added to the ConstraintFactory constructor.
- BackboneStub
- Constant
- ResidueType
- Rotamer
Other Function Types
These function types cannot currently be specified in a file. They need to have read_data methods implemented and be added to the FuncFactory constructor.
- PeriodicBound
- ChainbreakDist
- CharmmPeriodic
- CircularPower
- Etable
- CB_Angle
- CBSG_Dihedral
- SG_Dist
- SGSG_Dihedral
- LK_Sigmoidal