Results of Structure Prediction for Target Name: O53246 ( Click  )
)
Domain Boundary prediction ( View  )
)

Protein sequence
| 1-60: | M | A | G | A | K | H | A | G | R | I | V | A | I | T | T | A | A | A | V | I | L | A | A | C | S | S | G | S | K | G | G | A | G | S | G | H | A | G | K | A | R | S | A | V | T | T | T | D | A | D | W | K | P | V | A | D | A | L | G | R |
| 61-119: | S | G | K | L | G | D | N | N | T | A | Y | R | I | N | L | P | R | N | D | L | H | I | T | S | Y | G | V | D | I | K | P | G | L | S | L | G | G | Y | A | A | F | A | R | Y | D | N | N | E | T | L | L | M | G | D | L | V | I | T | E | E |
| 121-179: | E | L | P | K | V | T | D | A | L | Q | A | H | G | I | A | Q | T | A | L | H | K | H | L | L | Q | Q | D | P | P | V | W | W | T | H | I | H | G | M | G | D | A | A | R | L | A | Q | G | L | K | A | A | L | D | A | T | T | I | G | P | P |
| 181-239: | T | P | P | P | A | R | Q | P | P | V | D | I | D | V | A | G | V | D | Q | A | L | G | R | K | G | T | Q | D | G | G | L | M | K | Y | S | I | P | R | K | D | T | I | I | E | D | G | H | V | L | P | A | V | S | L | N | L | T | T | V | I |
| 241-299: | N | F | Q | P | V | G | R | G | R | A | A | I | N | G | D | F | I | L | I | A | P | E | V | Q | E | V | I | R | A | M | R | A | G | N | I | T | I | V | E | L | H | N | H | G | L | T | E | E | P | R | L | F | Y | M | H | Y | W | A | V | D |
| 301-321: | D | A | V | T | L | A | R | A | L | R | P | A | M | D | A | T | N | L | Q | S | S |
Secondary structure prediction (H: Helix E: Strand C: Coil)
| 1-60: | C | C | C | H | H | H | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | C | C |
| 61-119: | C | C | C | E | C | C | C | C | C | E | E | E | E | E | E | C | C | C | C | C | E | E | E | E | C | C | C | C | C | C | C | C | C | C | C | C | H | E | E | E | E | E | C | C | C | C | C | C | E | E | E | E | C | C | E | E | E | E | H | H |
| 121-179: | H | H | H | H | H | H | H | H | H | H | H | C | C | C | E | E | E | H | H | H | H | H | H | H | C | C | C | C | C | E | E | E | E | E | E | E | C | C | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C |
| 181-239: | C | C | C | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | E | E | E | E | E | E | C | C | C | C | C | E | E | E | C | C | C | C | C | C | C | C | C | C | C | C | E | E | E | E |
| 241-299: | E | E | C | C | C | C | C | C | E | E | E | E | C | C | C | E | E | E | E | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | E | E | E | E | E | H | H | H | H | C | C | C | C | C | C | E | E | E | E | E | E | E | C | C | C |
| 301-321: | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C |
| H(Helix): 103(32.09%) | E(Strand): 69(21.5%) | C(Coil): 149(46.42%) |
Solvent accessibility prediction (e: Exposed b: Buried)
| 1-60: | E | E | B | B | E | E | B | E | E | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | E | B | E | E | E | E | E | E | E | E | E | B | E | E | B | E | E | E | E | E | E | E | E | B | E | B | E | E | B | E | E | B | B | E | E |
| 61-119: | E | B | E | B | E | E | E | E | B | B | B | B | B | E | B | E | B | E | E | B | E | B | E | B | E | E | E | E | B | E | B | E | B | B | B | B | B | B | B | B | B | B | E | B | E | E | E | E | B | B | B | B | B | B | B | B | B | B | B | E |
| 121-179: | B | B | E | E | B | B | E | B | B | E | E | E | E | B | E | B | B | B | B | B | B | B | B | B | E | E | E | B | B | B | B | B | B | B | B | B | B | E | E | E | B | B | E | B | B | E | E | B | E | E | B | B | E | E | B | E | E | E | E | E |
| 181-239: | E | E | E | E | E | E | E | E | E | E | E | B | E | B | E | E | B | B | E | B | B | E | E | E | B | E | B | E | E | B | B | B | B | B | B | B | E | B | E | E | E | B | B | E | E | E | E | E | B | B | E | E | E | B | B | B | B | B | B | B |
| 241-299: | B | B | B | B | B | E | E | E | E | B | B | B | B | B | B | B | B | B | B | B | E | E | B | E | E | B | B | E | B | B | E | E | E | E | B | E | B | B | B | B | B | B | B | B | B | E | E | E | B | B | B | B | B | B | B | B | B | B | E | E |
| 301-321: | E | B | E | E | B | B | E | E | B | E | E | B | B | E | E | E | E | E | E | E | E |
| e(Exposed): 150(46.73%) | b(Buried): 171(53.27%) |
Disorder prediction (N: Normal T: Disorder)
| 1-60: | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 61-119: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 121-179: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 181-239: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 241-299: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 301-321: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T |
| N(Normal): 287(89.41%) | T(Disorder): 34(10.59%) |
Select domain
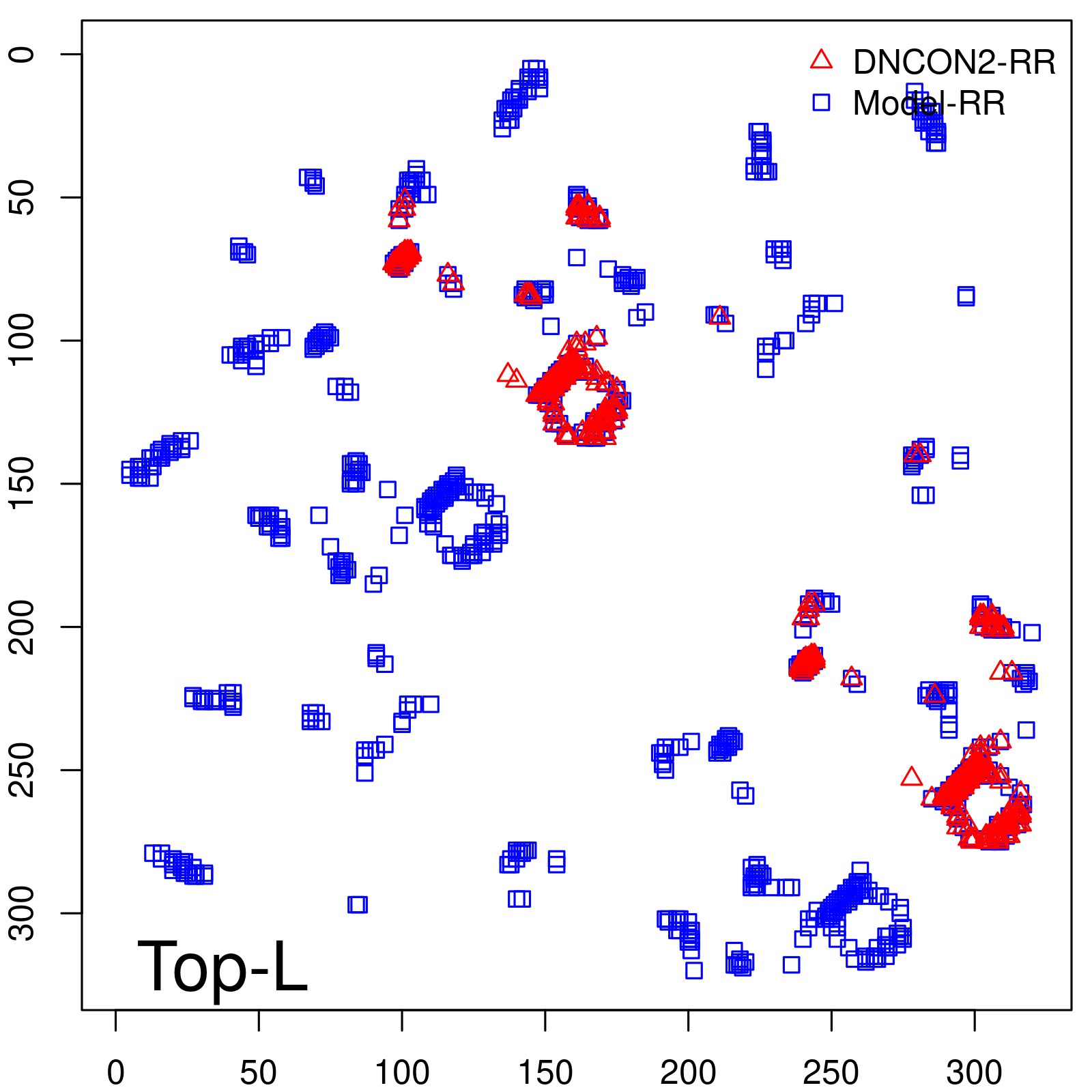
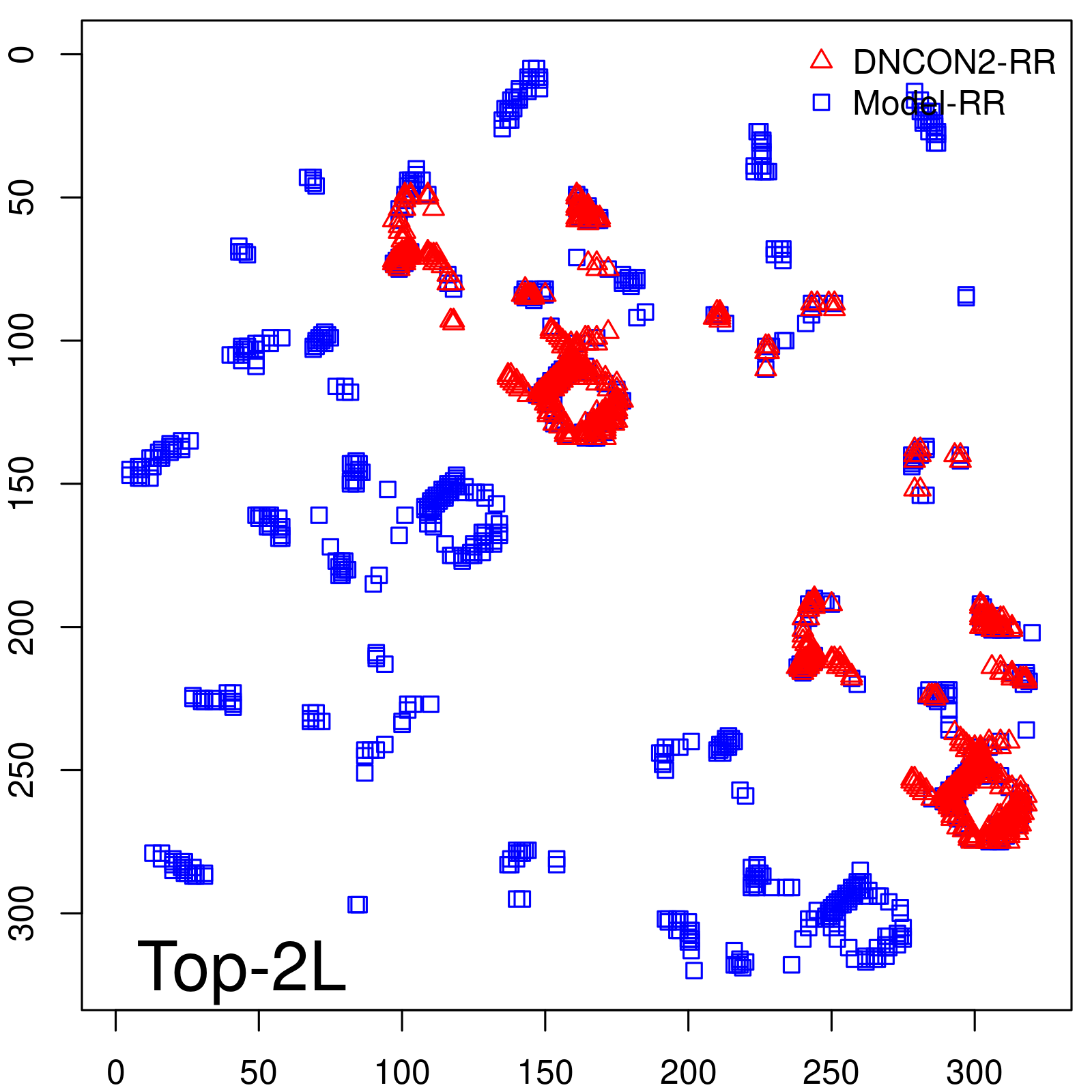
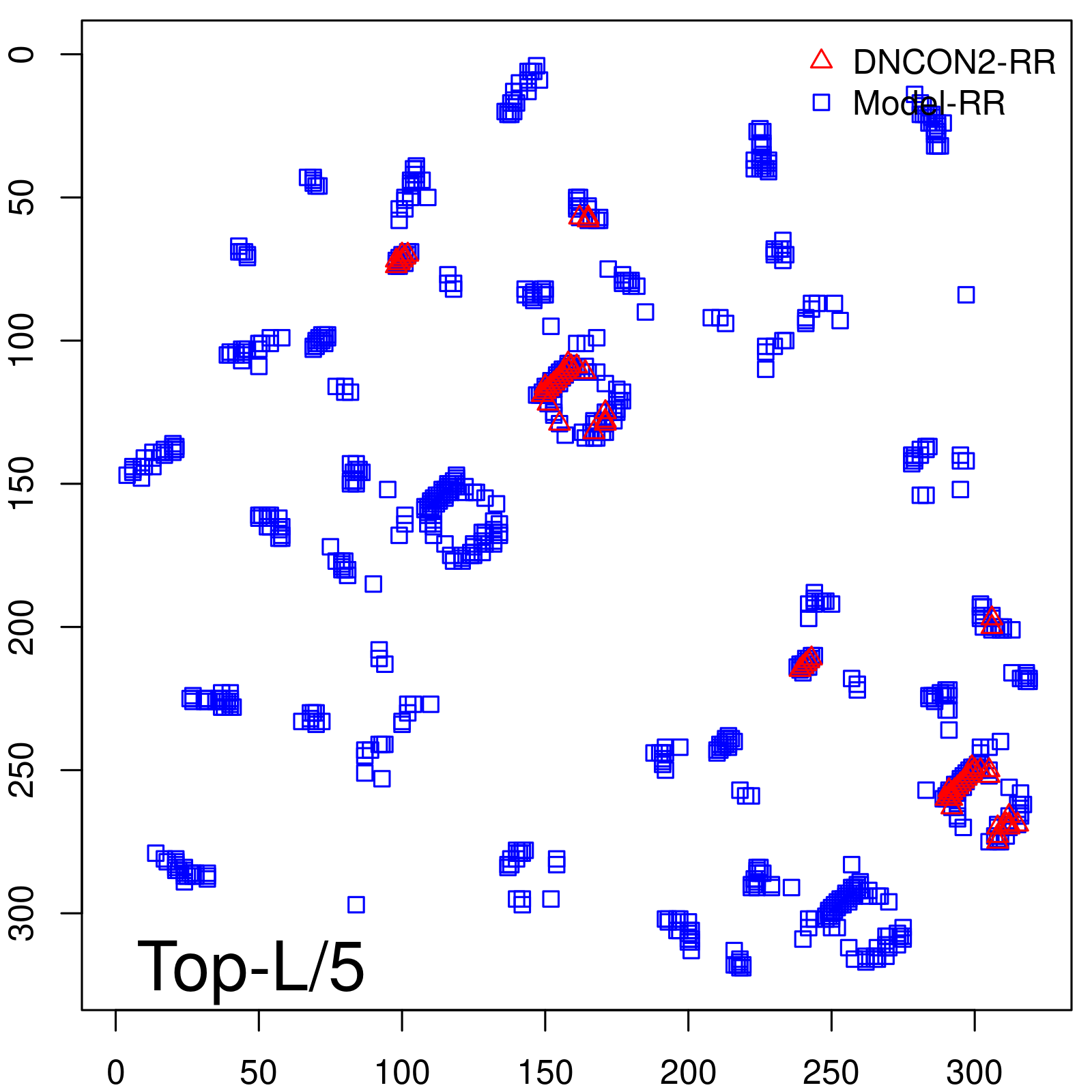
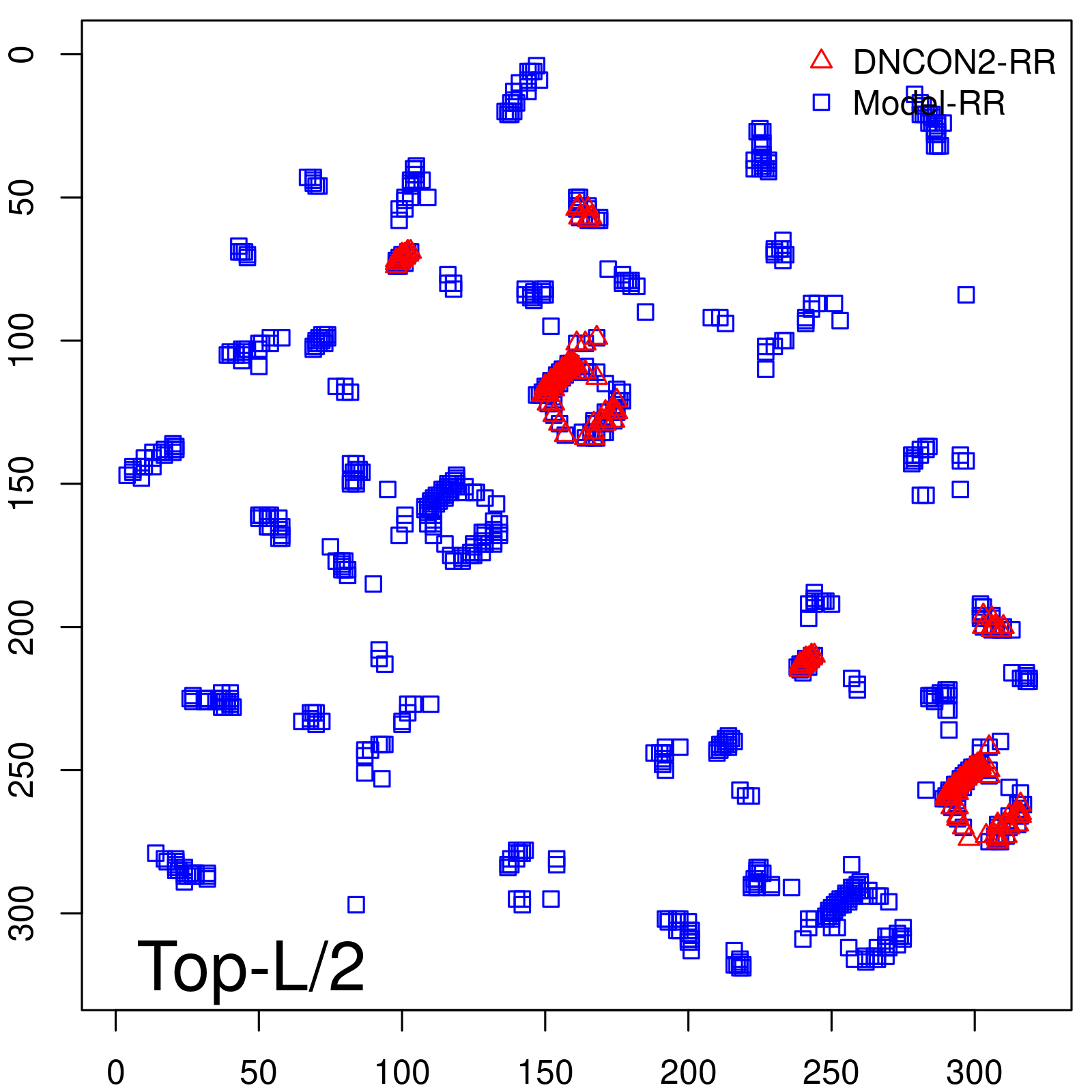
Predicted contact map and distance map
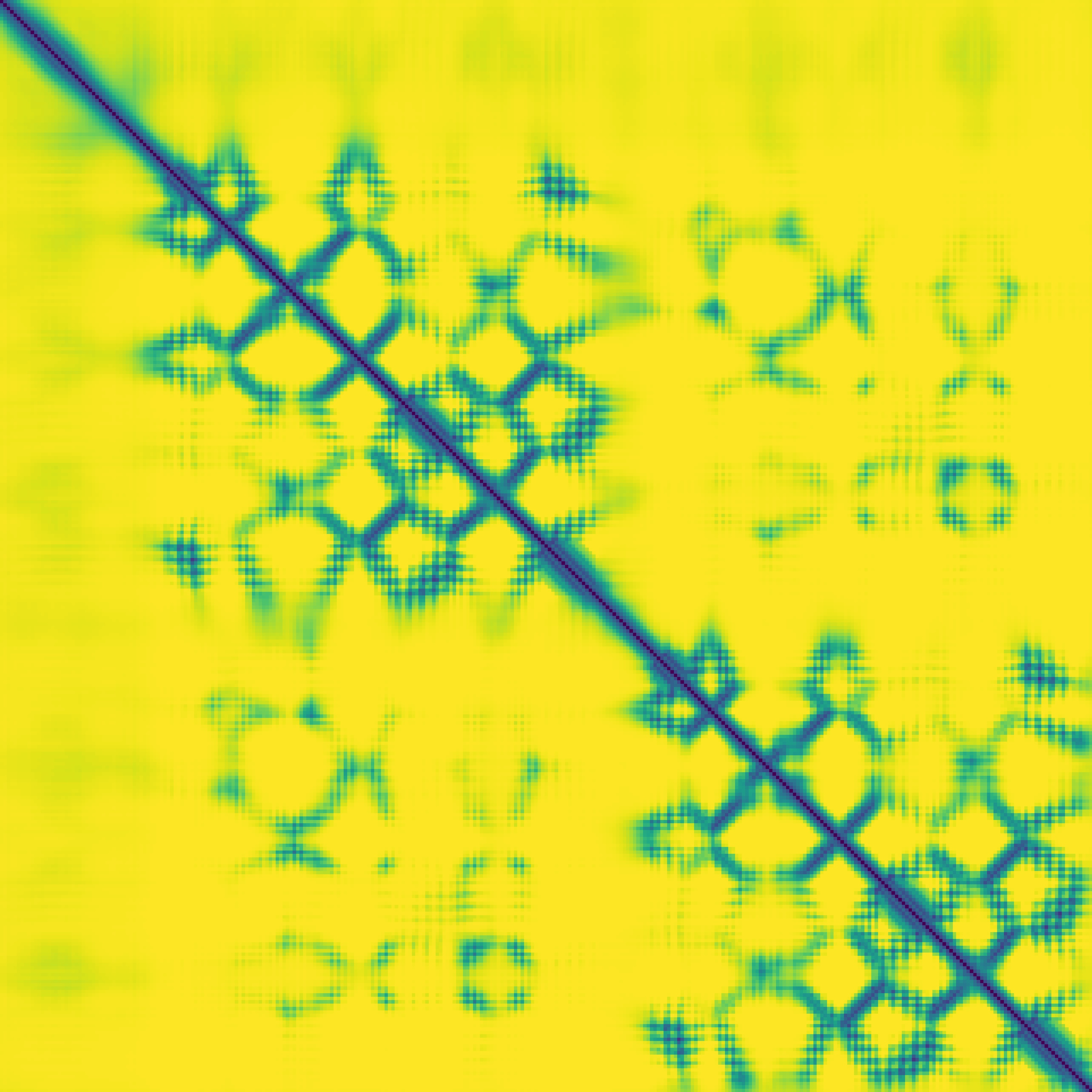
|
|

|
Probability to Precision
|
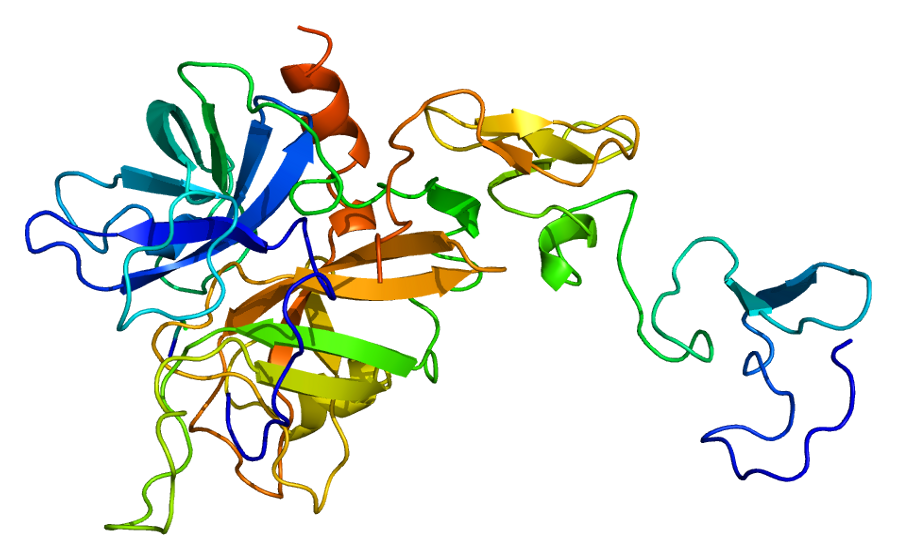
Predicted Top 1 Tertiary structure
|
|
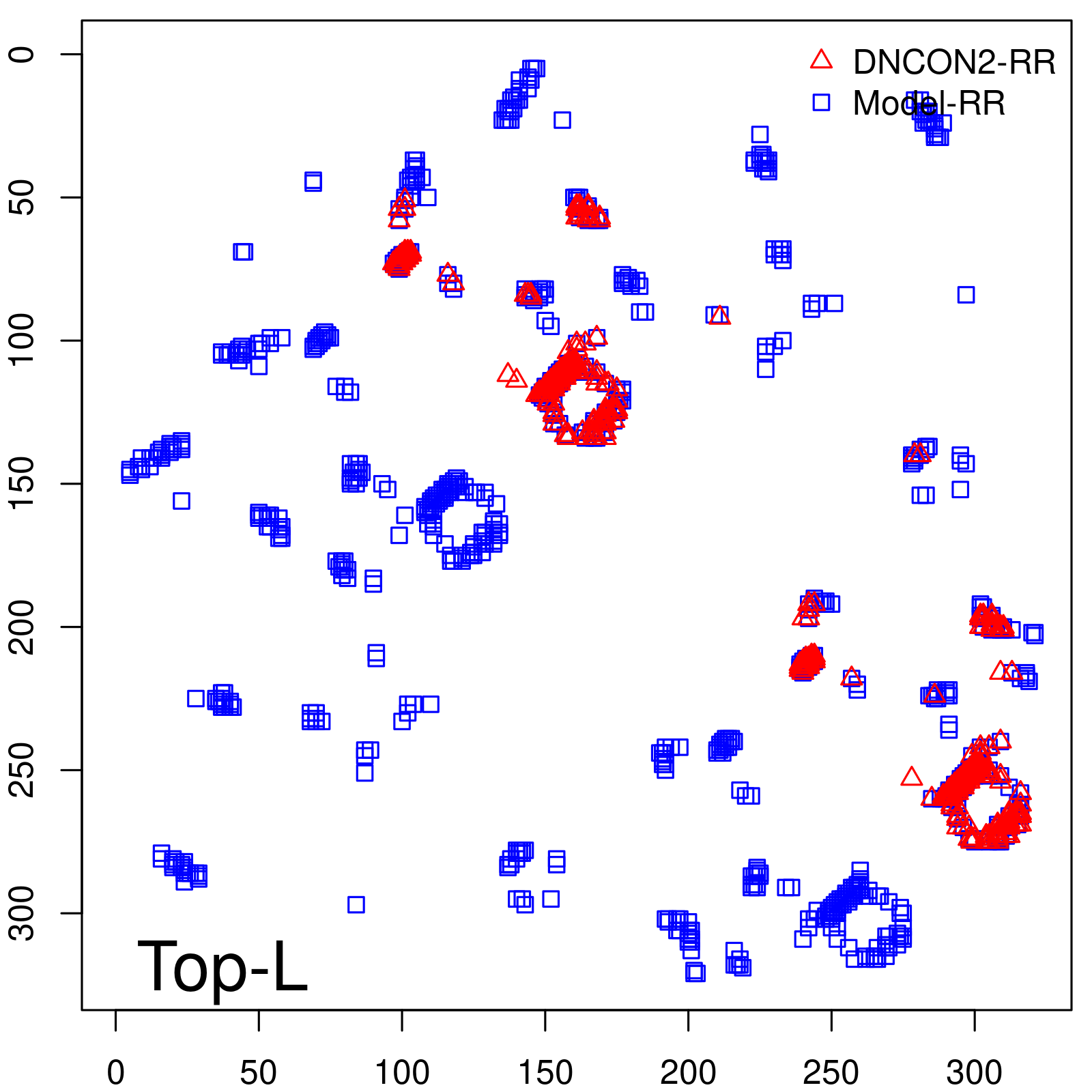
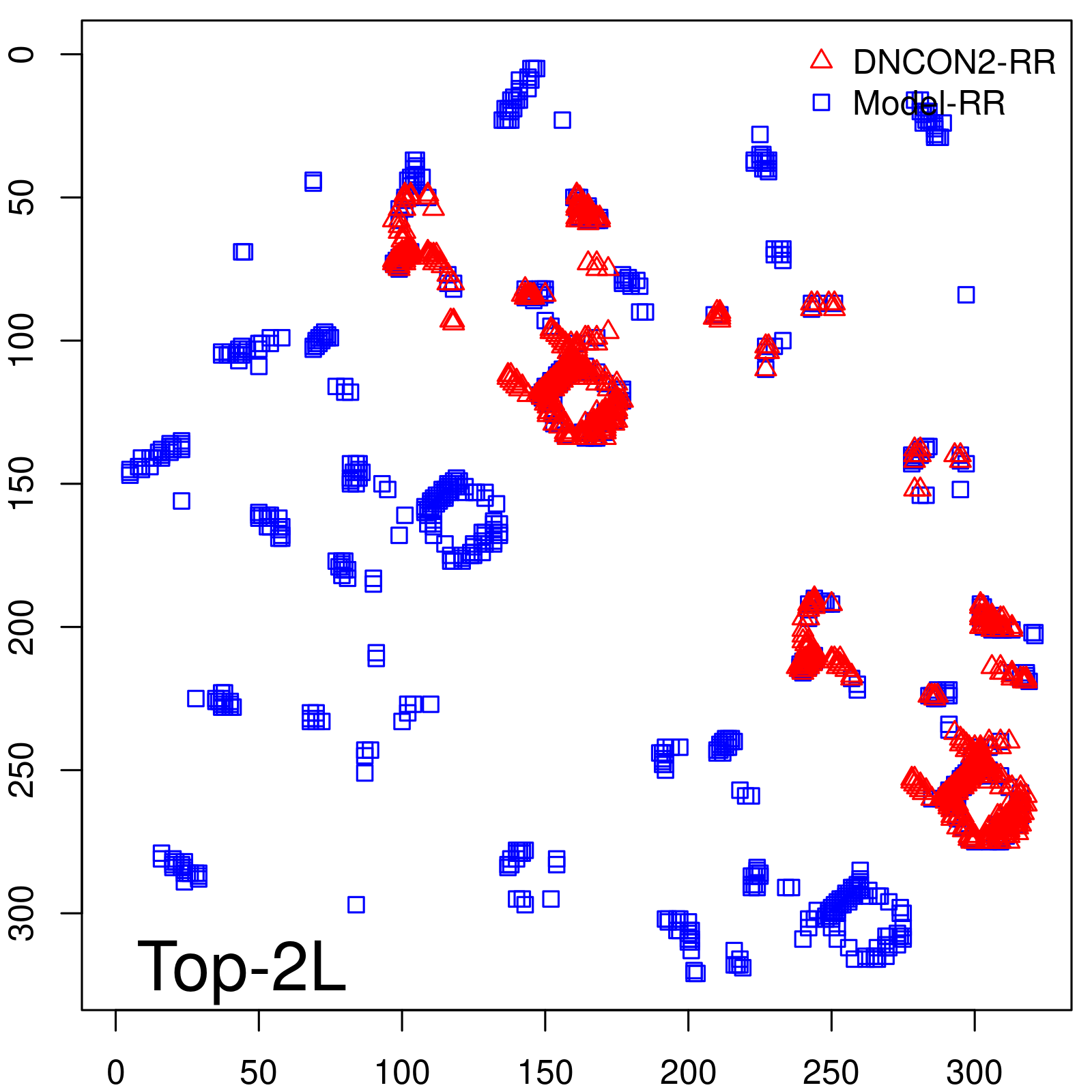
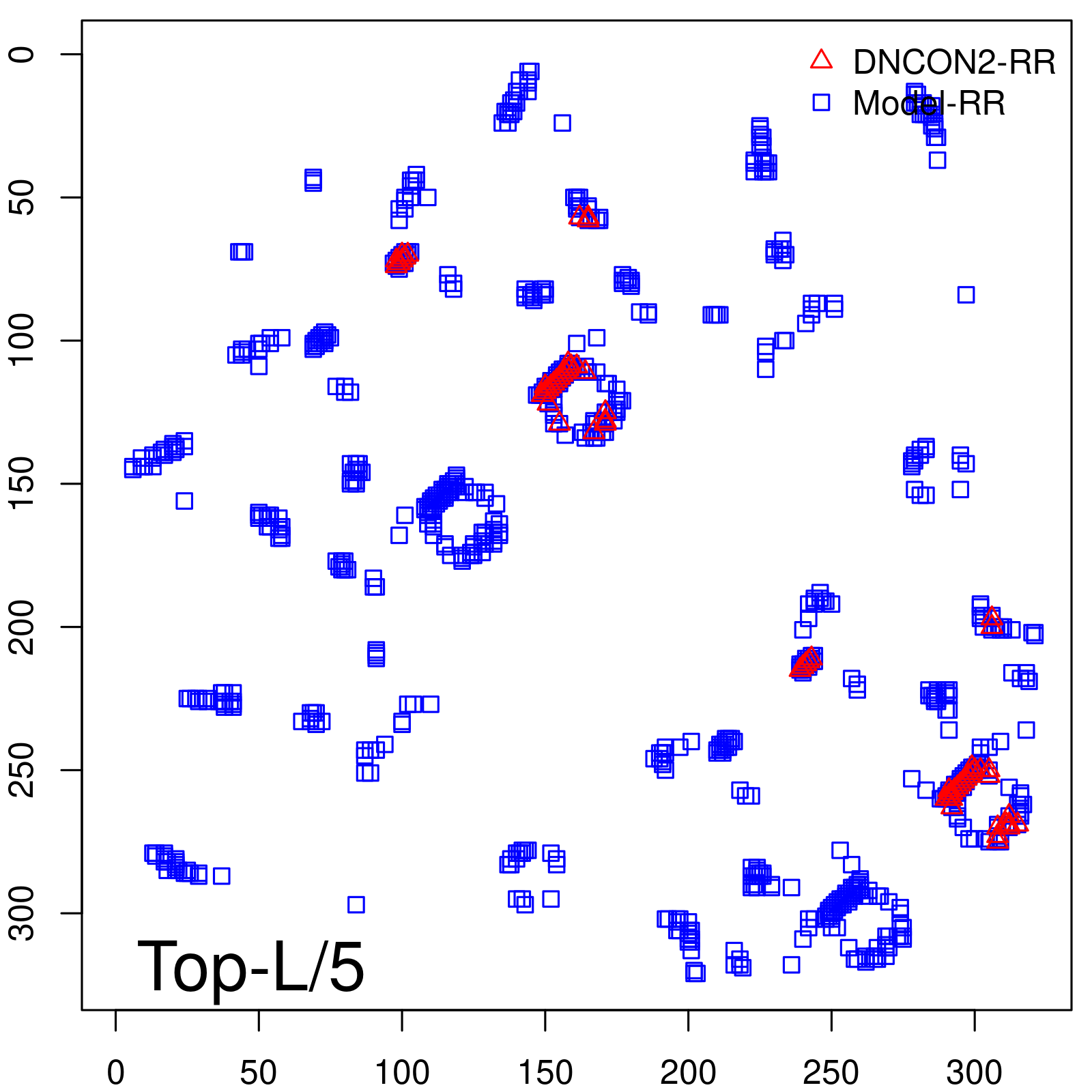
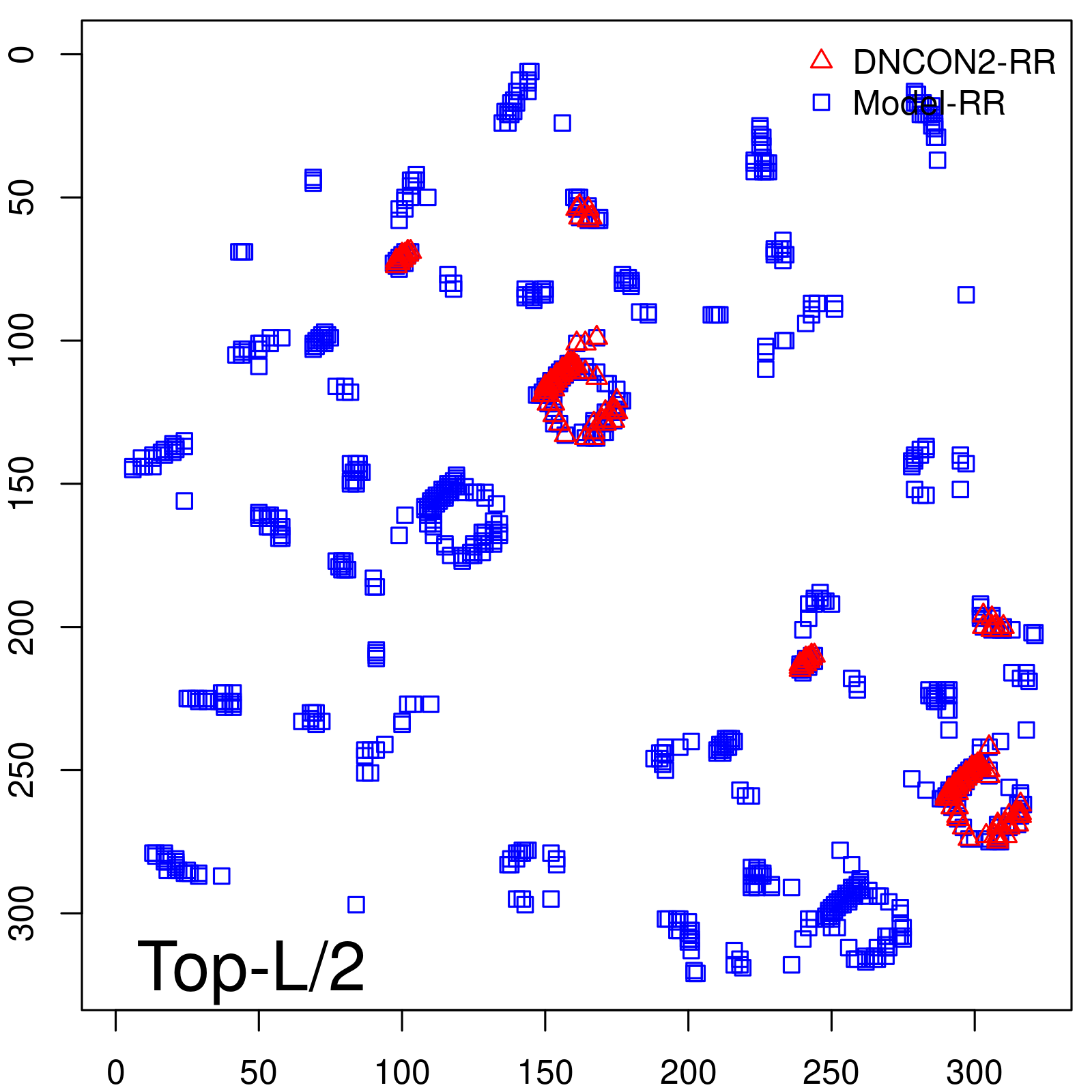
Predicted Contact Accuracy
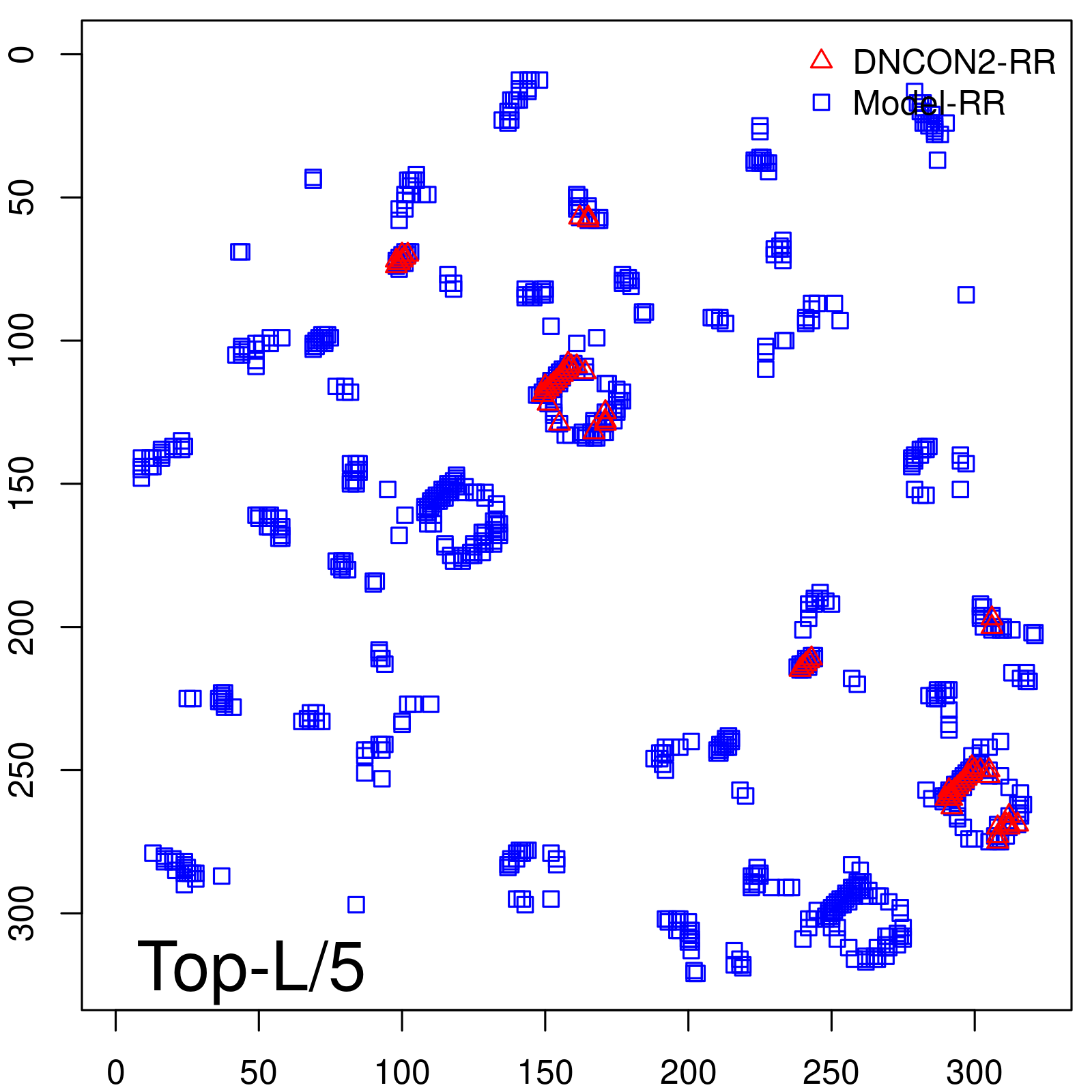
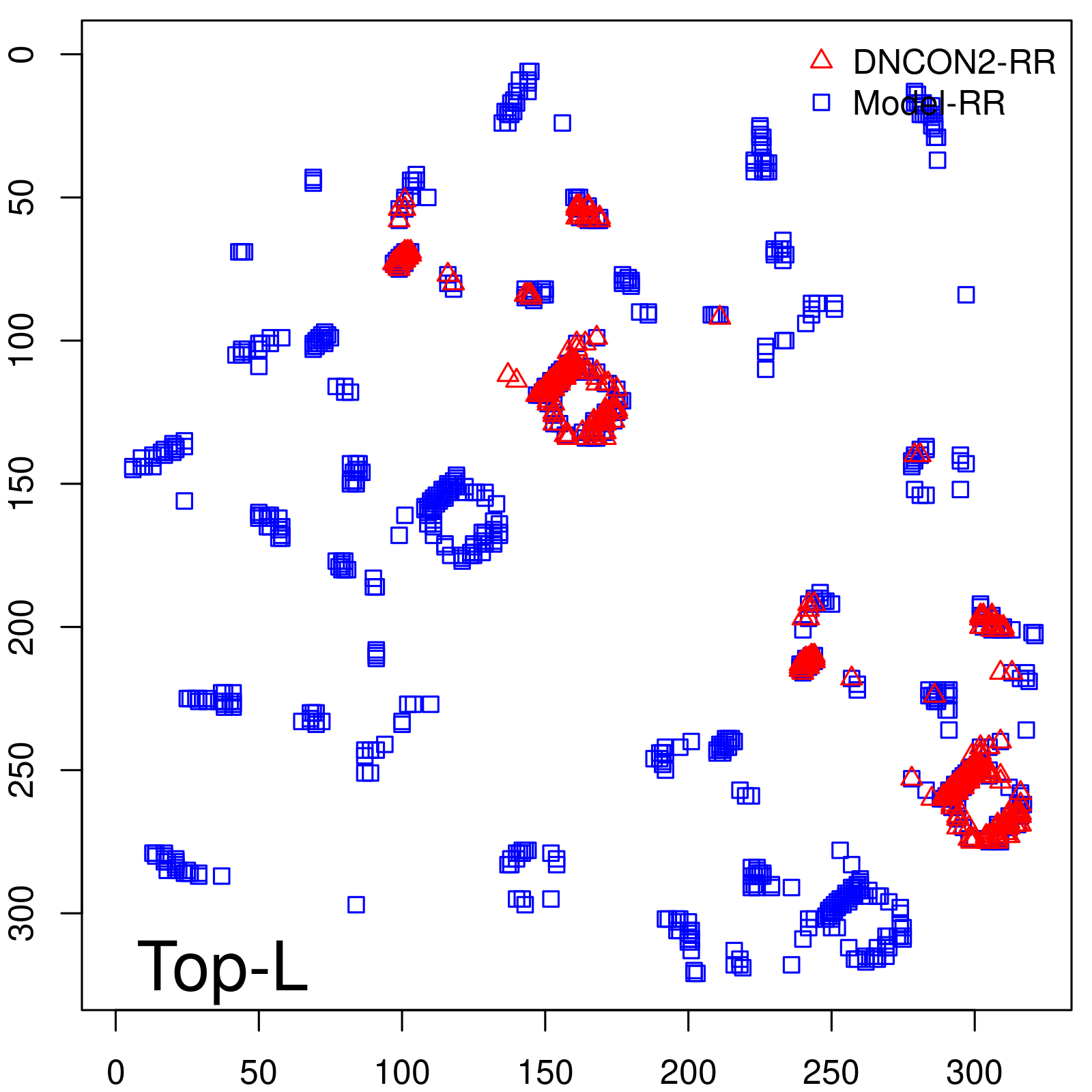
|
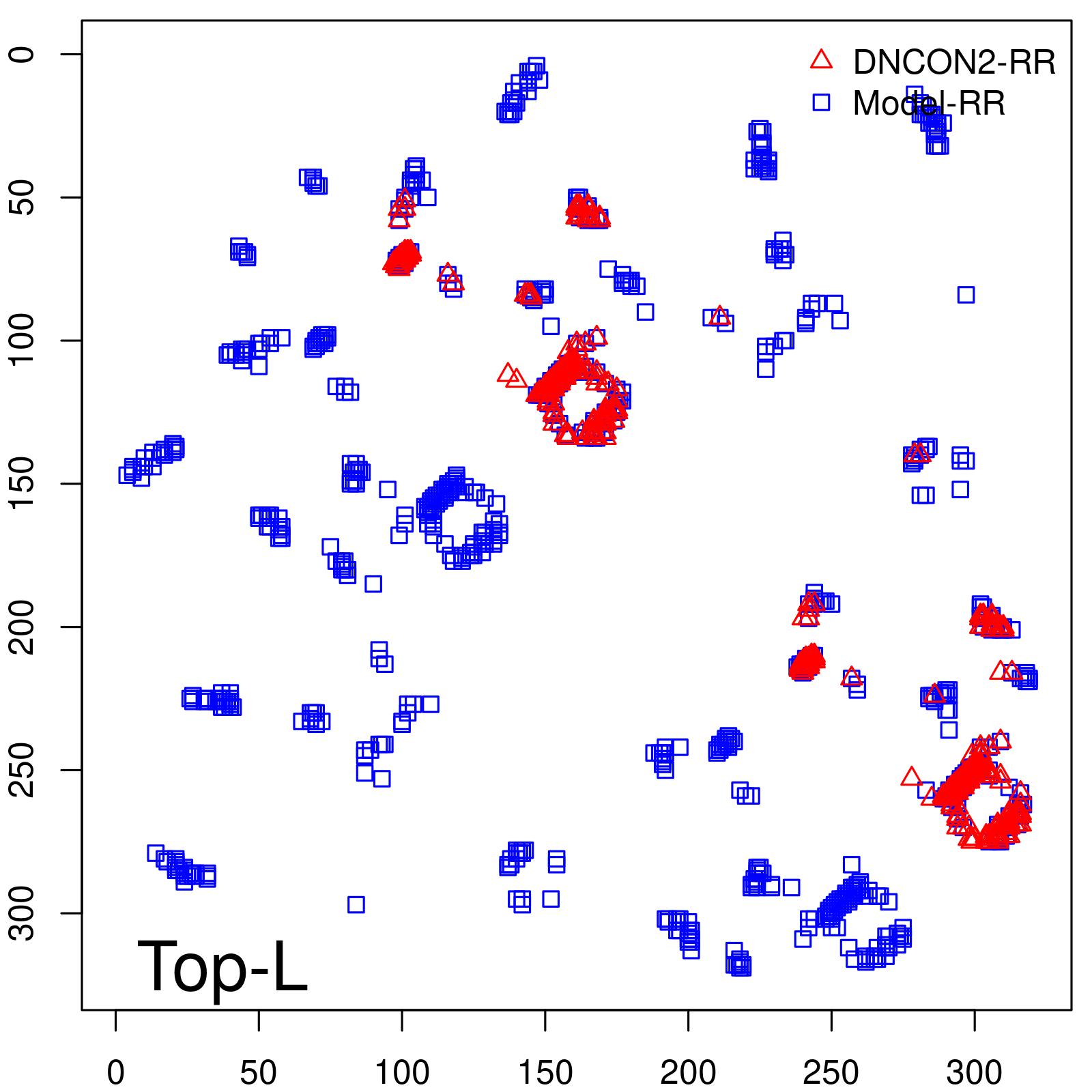
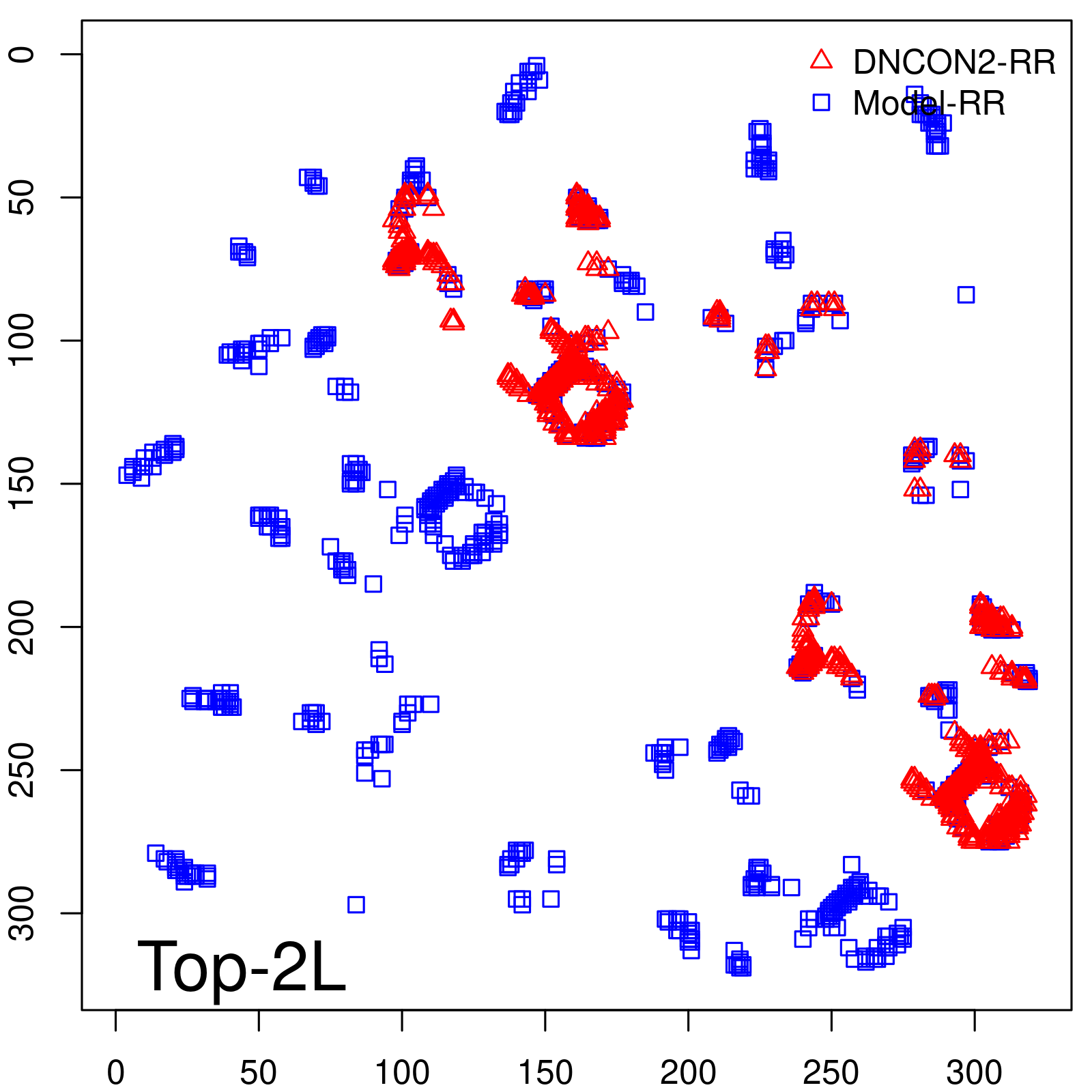
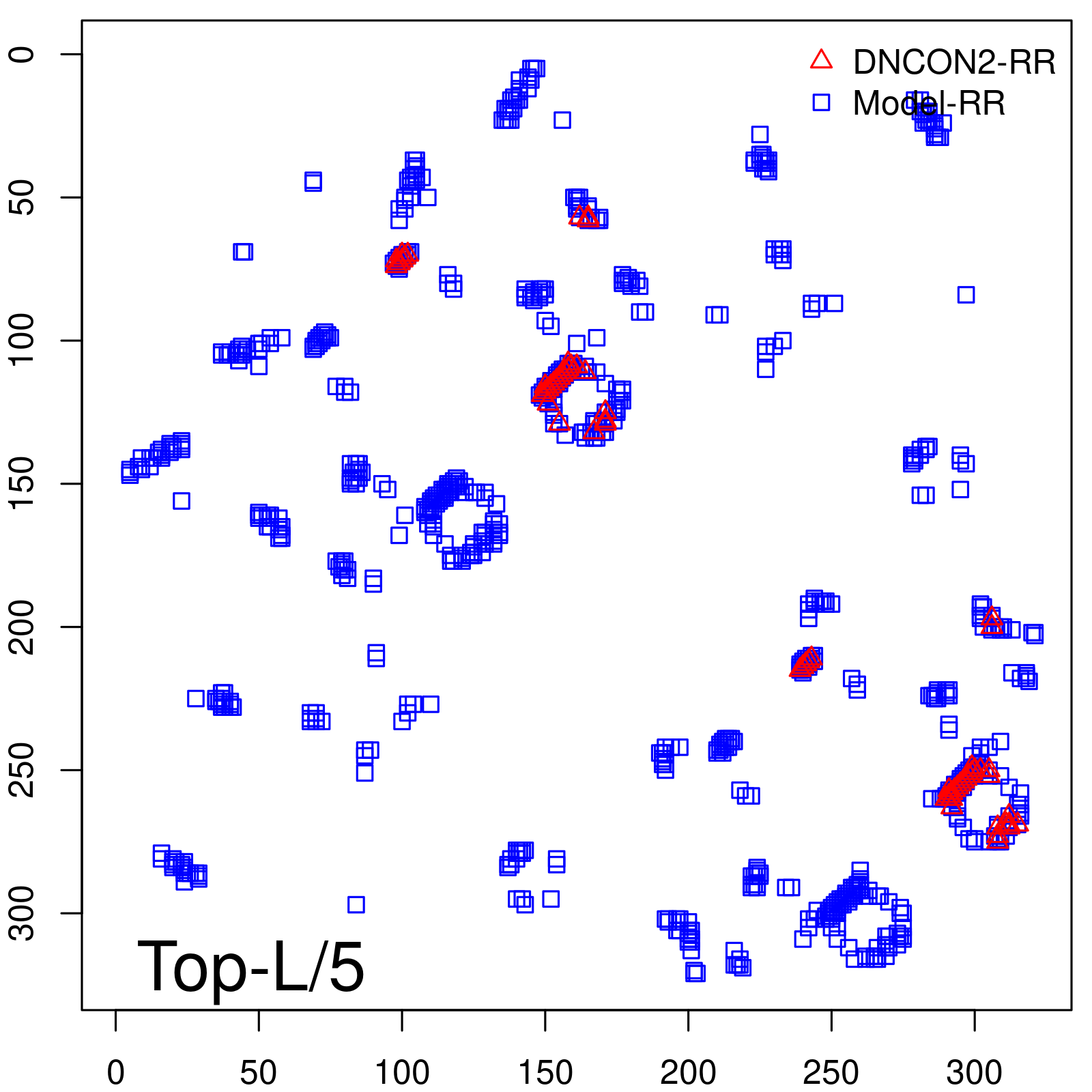
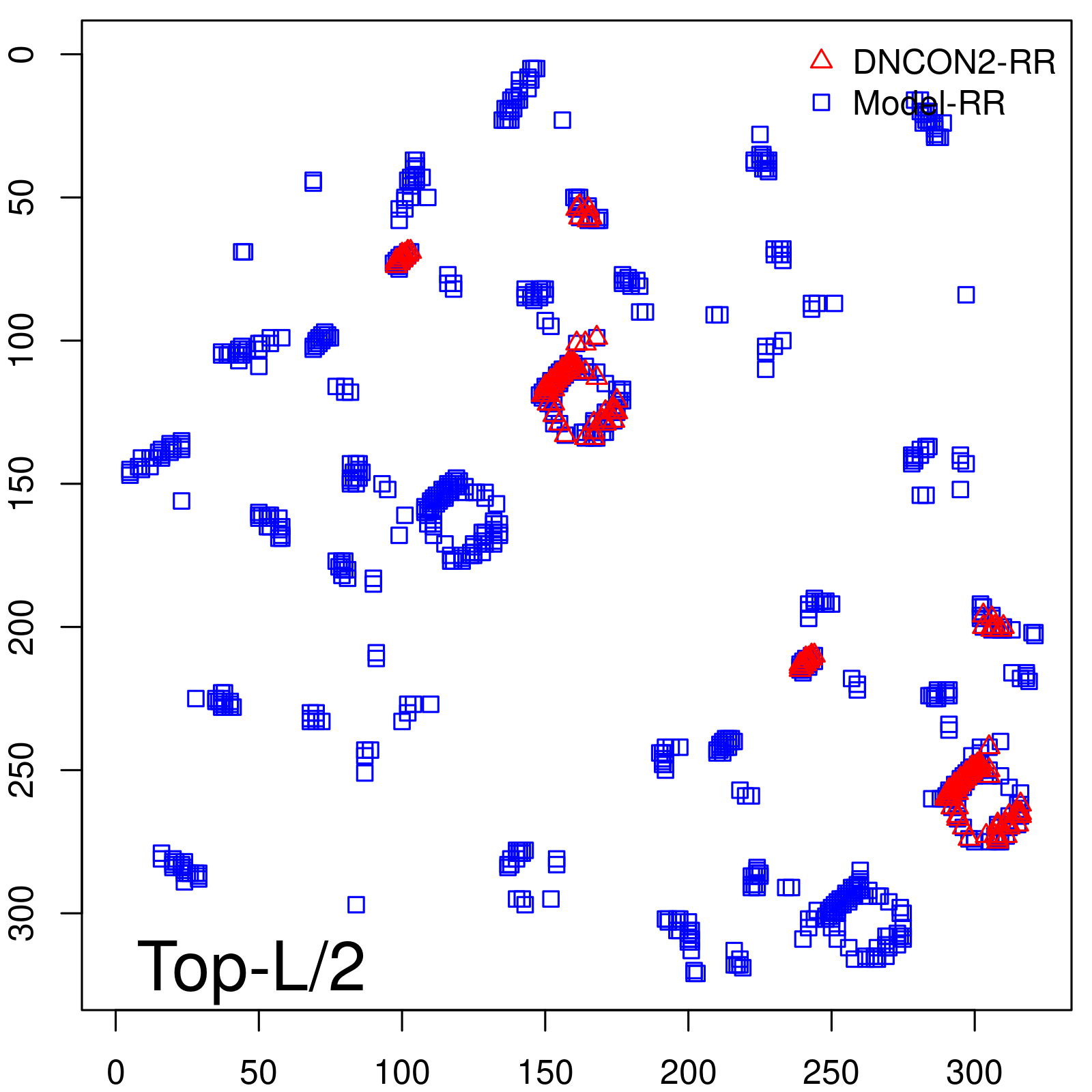
| Long-Range | Precision |
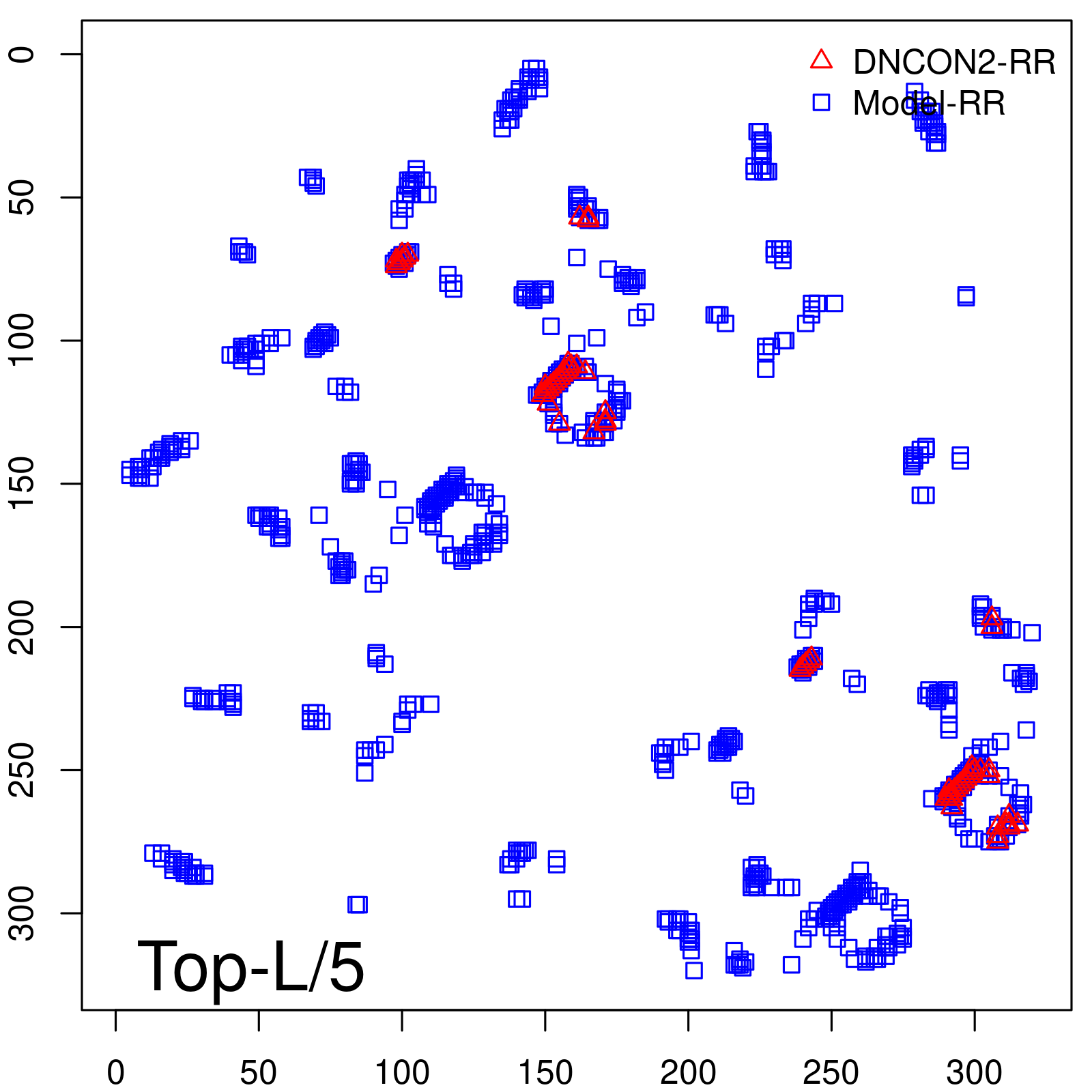
| TopL/5 | 100.00 |
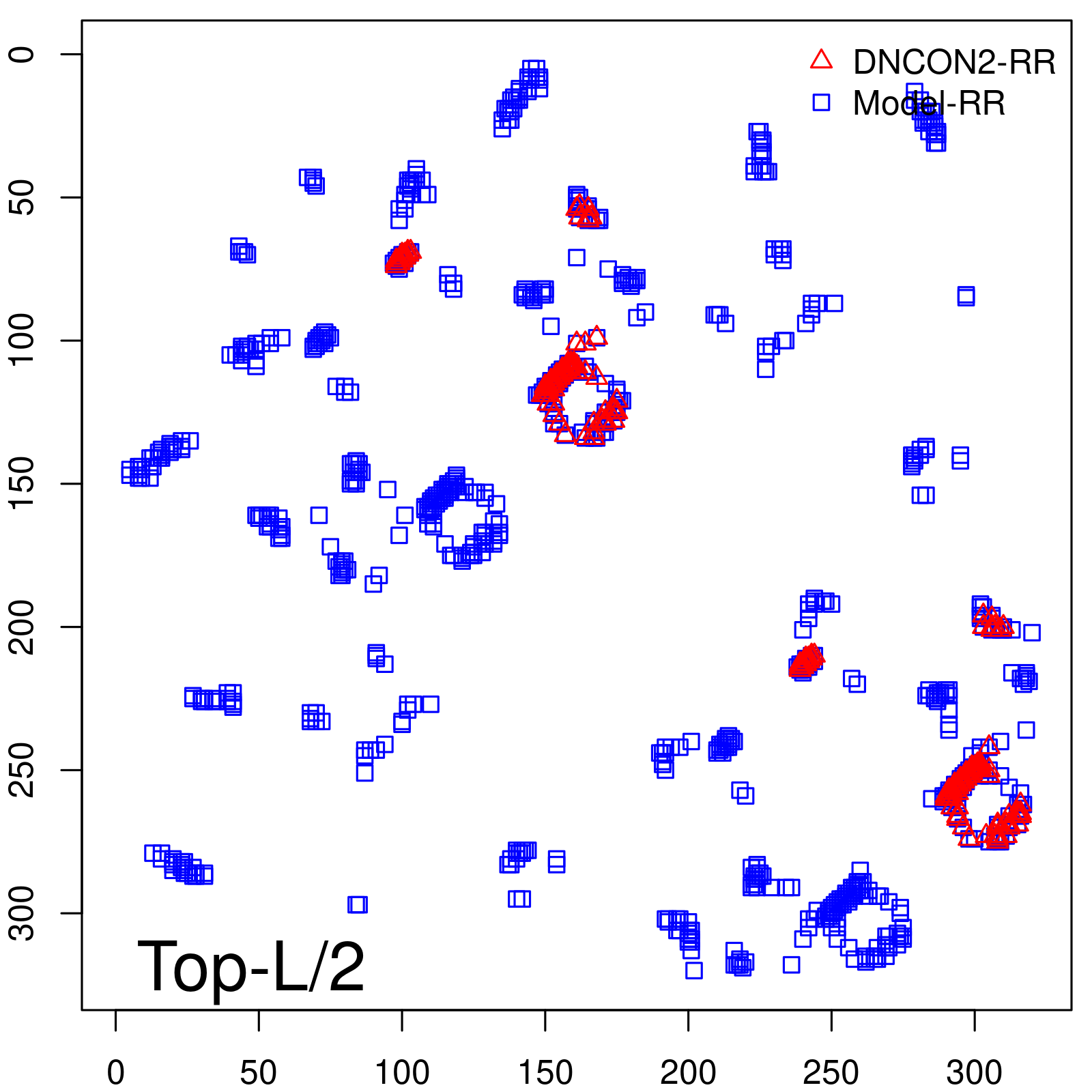
| TopL/2 | 96.89 |
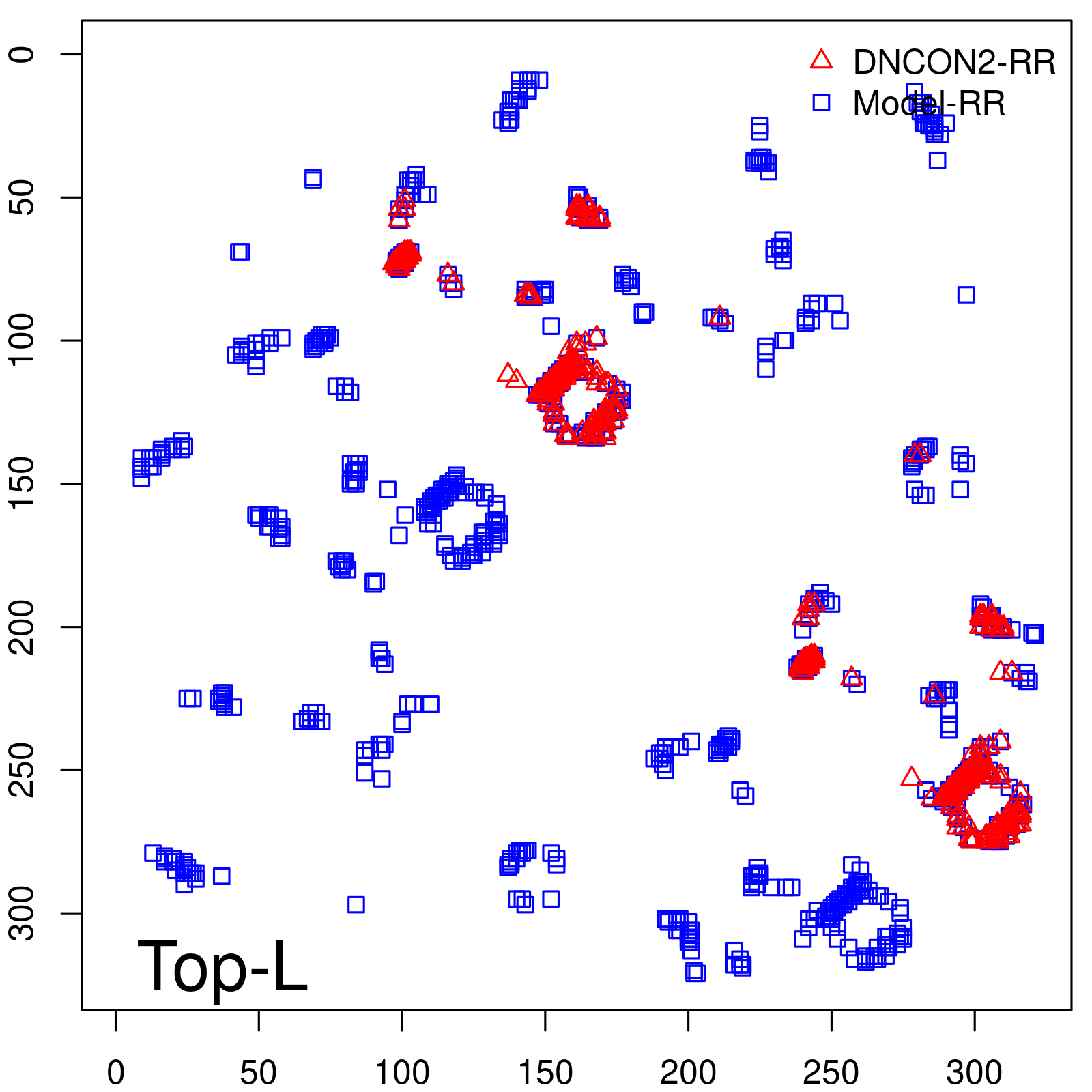
| TopL | 73.21 |
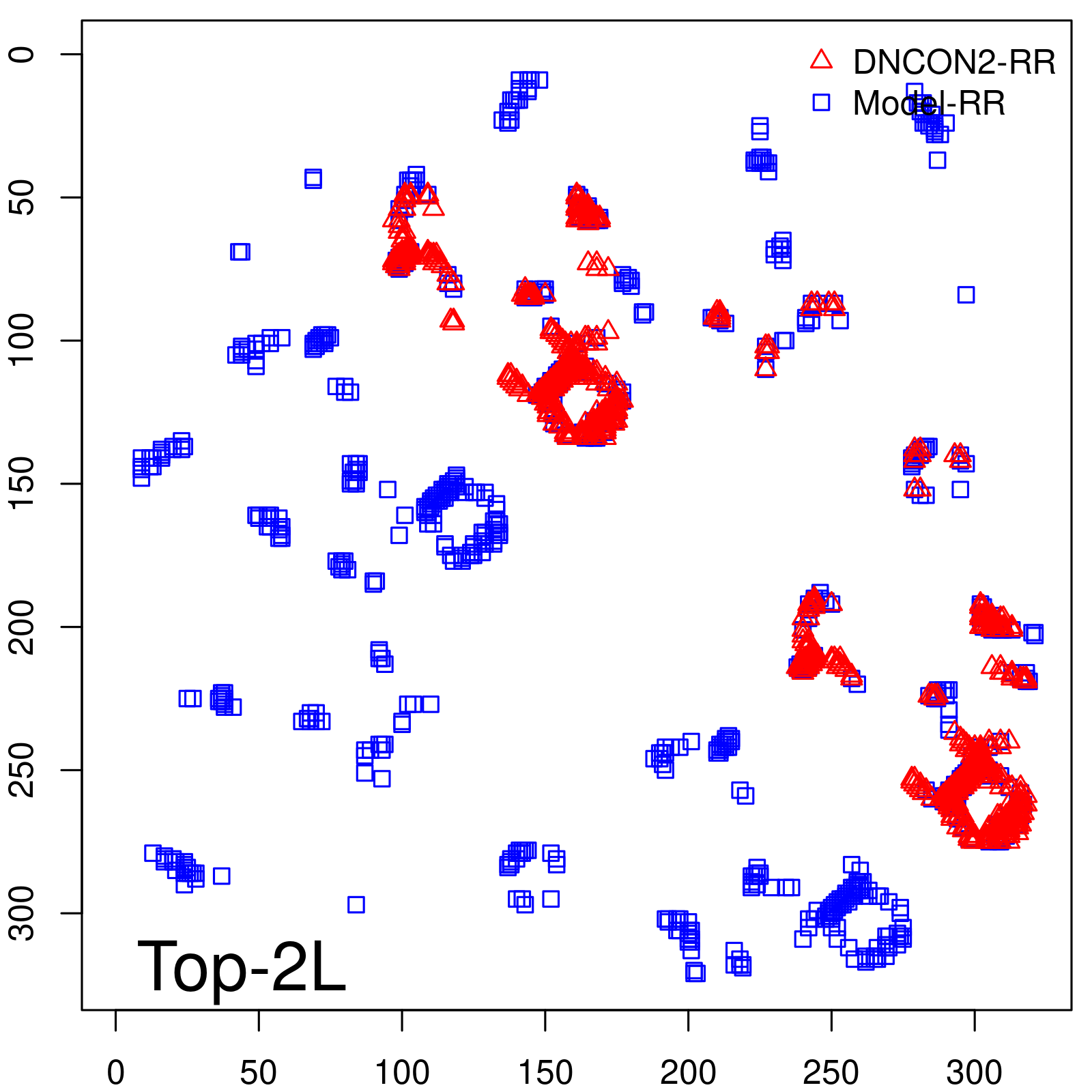
| Top2L | 44.86 |
( Alignment file  )
)
| Alignment | Number |
| N | 1479 |
| Neff | 137 |
Model comparsion
| Model | TM-score |
| Model 2 | 0.9529 |
| Model 3 | 0.9517 |
| Model 4 | 0.9495 |
| Model 5 | 0.9534 |
| Average | 0.95188 |
Radius Gyration
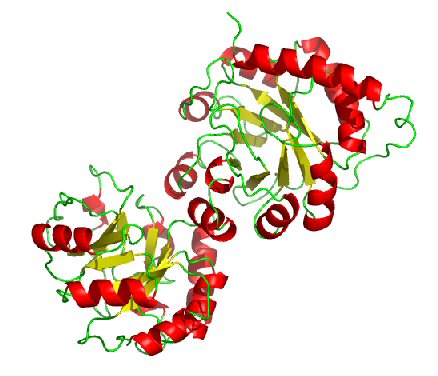
Similar pdbs
Predicted Top 2 Tertiary structure
|
|
Predicted Contact Accuracy
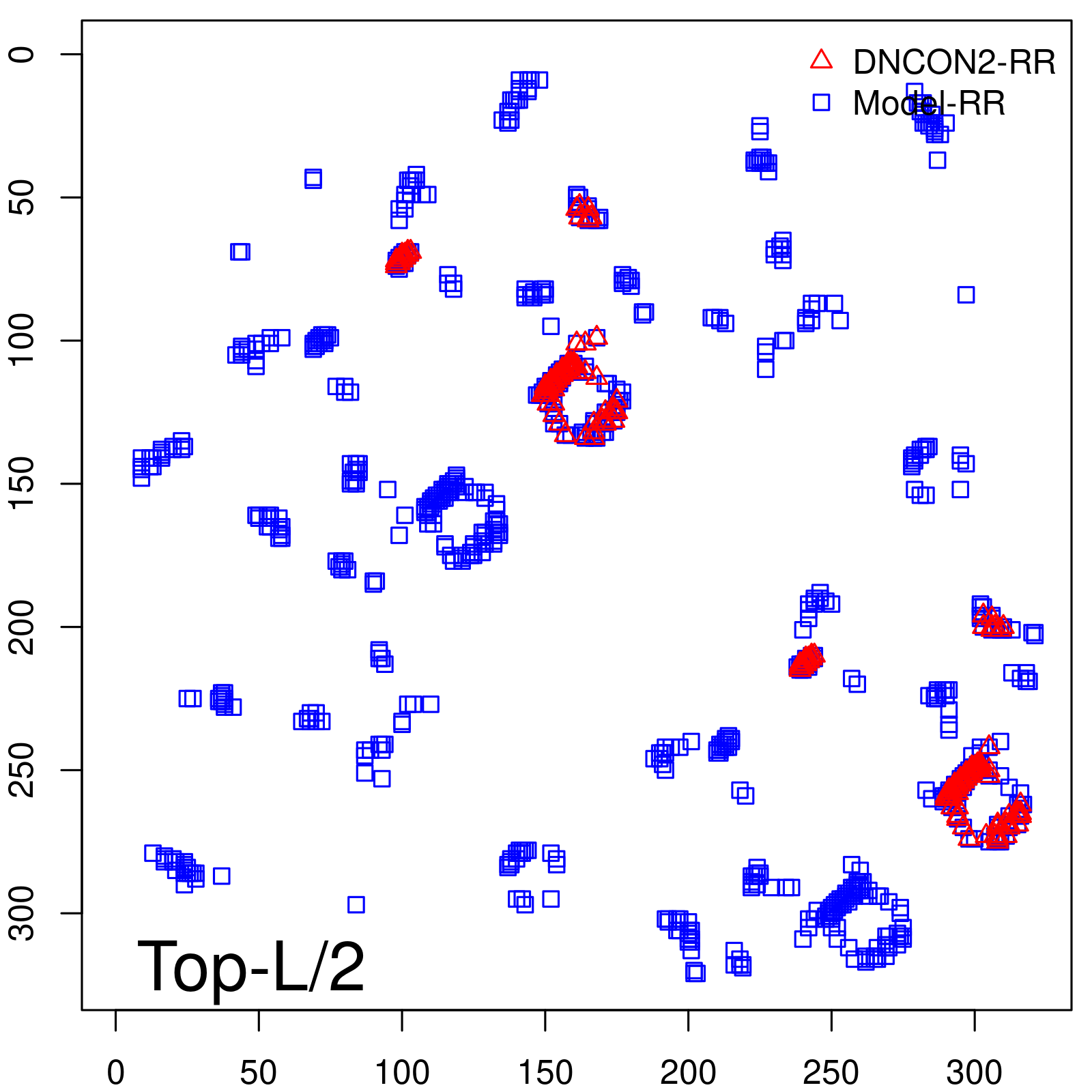
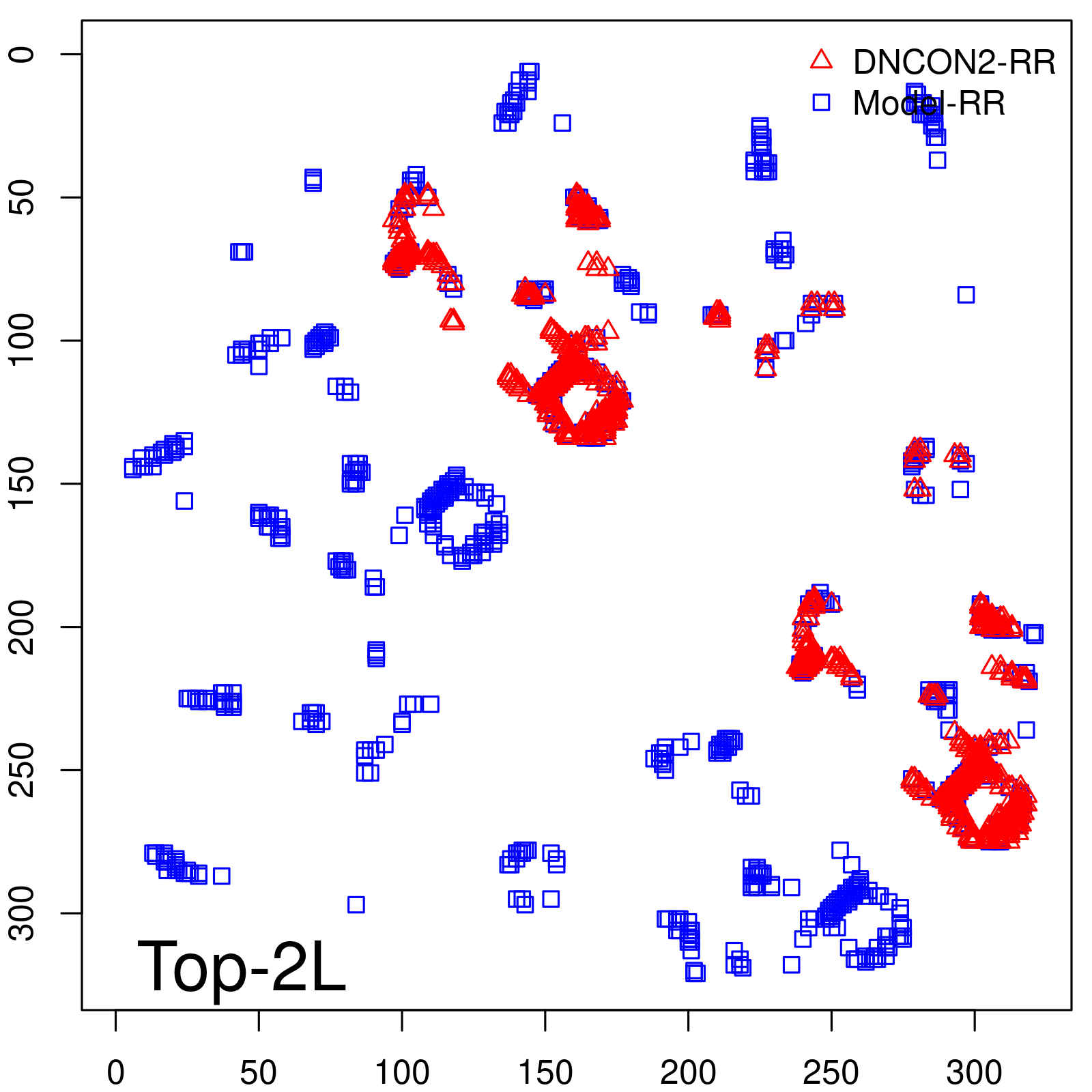
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 95.65 |
| TopL | 74.14 |
| Top2L | 44.86 |
( Alignment file  )
)
| Alignment | Number |
| N | 1479 |
| Neff | 137 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9529 |
| Model 3 | 0.9478 |
| Model 4 | 0.9550 |
| Model 5 | 0.9345 |
| Average | 0.94755 |
Radius Gyration
Similar pdbs
Predicted Top 3 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 96.27 |
| TopL | 71.34 |
| Top2L | 42.83 |
( Alignment file  )
)
| Alignment | Number |
| N | 1479 |
| Neff | 137 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9517 |
| Model 2 | 0.9478 |
| Model 4 | 0.9484 |
| Model 5 | 0.9501 |
| Average | 0.94950 |
Radius Gyration
Similar pdbs
Predicted Top 4 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 96.89 |
| TopL | 75.08 |
| Top2L | 44.70 |
( Alignment file  )
)
| Alignment | Number |
| N | 1479 |
| Neff | 137 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9495 |
| Model 2 | 0.9550 |
| Model 3 | 0.9484 |
| Model 5 | 0.9364 |
| Average | 0.94732 |
Radius Gyration
Similar pdbs
Predicted Top 5 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 95.65 |
| TopL | 74.77 |
| Top2L | 45.02 |
( Alignment file  )
)
| Alignment | Number |
| N | 1479 |
| Neff | 137 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9534 |
| Model 2 | 0.9345 |
| Model 3 | 0.9501 |
| Model 4 | 0.9364 |
| Average | 0.94360 |
Radius Gyration
Similar pdbs