P64524
multicom
P64524
full_length
P64524
Results of Structure Prediction for Target Name: P64524 ( Click  )
)
Domain Boundary prediction ( View  )
)

>P64524: 1-124
| 1-60: |
M | K | T | L | P | V | L | P | G | Q | A | A | S | S | R | P | S | P | V | E | I | W | Q | I | L | L | S | R | L | L | D | Q | H | Y | G | L | T | L | N | D | T | P | F | A | D | E | R | V | I | E | Q | H | I | E | A | G | I | S | L | C |
| 61-119: |
D | A | V | N | F | L | V | E | K | Y | A | L | V | R | T | D | Q | P | G | F | S | A | C | T | R | S | Q | L | I | N | S | I | D | I | L | R | A | R | R | A | T | G | L | M | T | R | D | N | Y | R | T | V | N | N | I | T | L | G | K | Y |
| 121-124: |
P | E | A | K |
| 1-60: |
C | C | C | C | C | C | C | H | H | H | H | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | E | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H | C | C | C | C | H | H |
| 61-119: |
H | H | H | H | H | H | H | H | H | H | C | E | E | E | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | E | E | E | C | C | E | C | C | C | C | C |
| 121-124: |
C | C | C | C |
|
| | H(Helix): 51(41.13%) | E(Strand): 8(6.45%) | C(Coil): 65(52.42%) |
| 1-60: |
E | E | E | B | E | E | E | E | E | E | B | B | E | E | B | E | B | B | B | B | B | B | B | B | B | B | B | E | B | B | E | E | B | B | B | B | B | B | E | E | B | E | B | B | E | E | E | B | B | E | E | B | B | E | E | E | B | E | B | B |
| 61-119: |
E | B | B | B | E | B | B | E | E | B | E | B | E | B | B | E | E | E | E | B | E | B | B | E | E | B | E | B | B | B | B | B | B | B | B | B | B | B | E | B | B | B | B | B | E | E | E | E | B | E | E | B | E | E | B | E | E | E | E | E |
| 121-124: |
E | E | E | E |
|
| | e(Exposed): 59(47.58%) | b(Buried): 65(52.42%) |
| 1-60: |
T | T | T | T | T | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 61-119: |
N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T |
| 121-124: |
T | T | T | T |
|
| | N(Normal): 107(86.29%) | T(Disorder): 17(13.71%) |
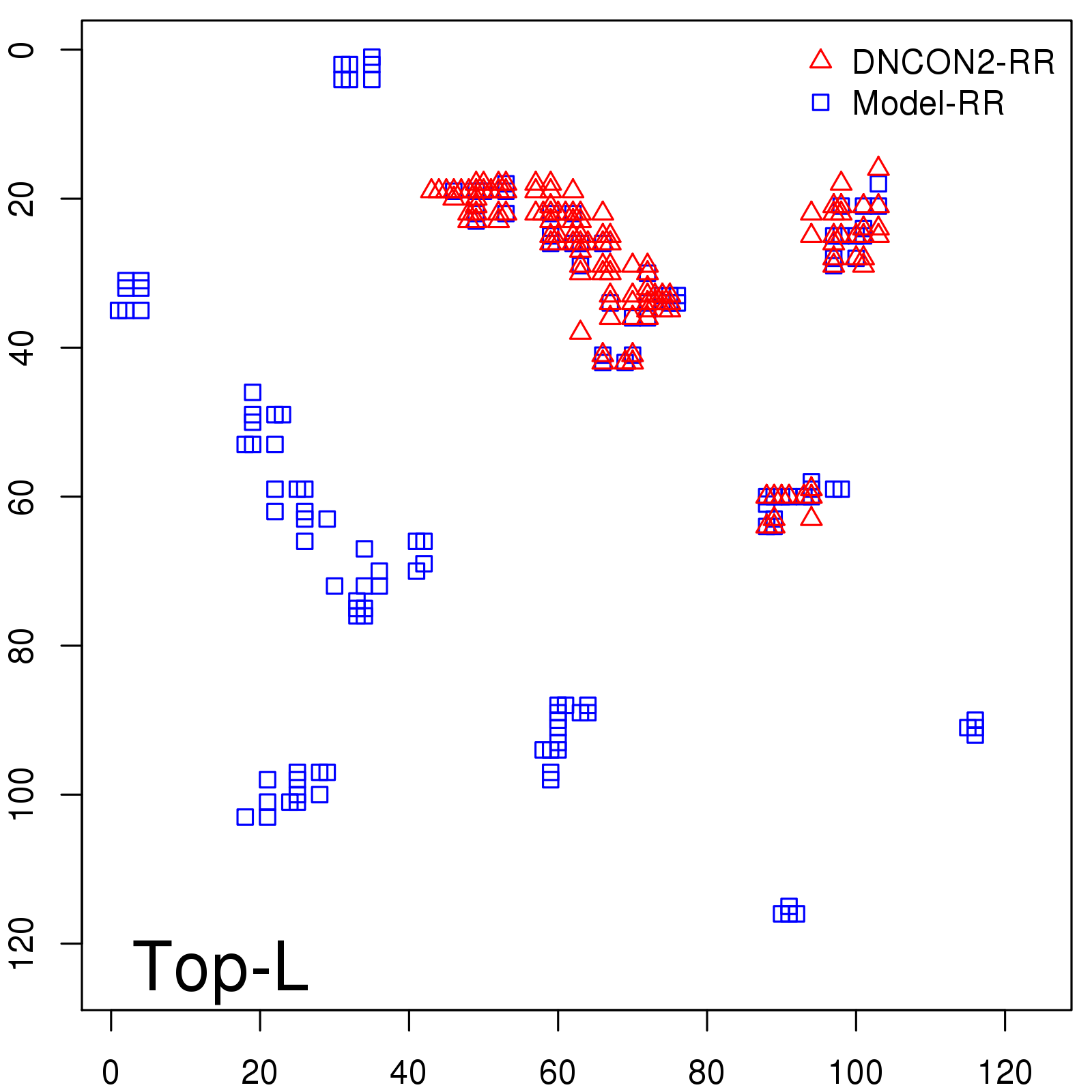
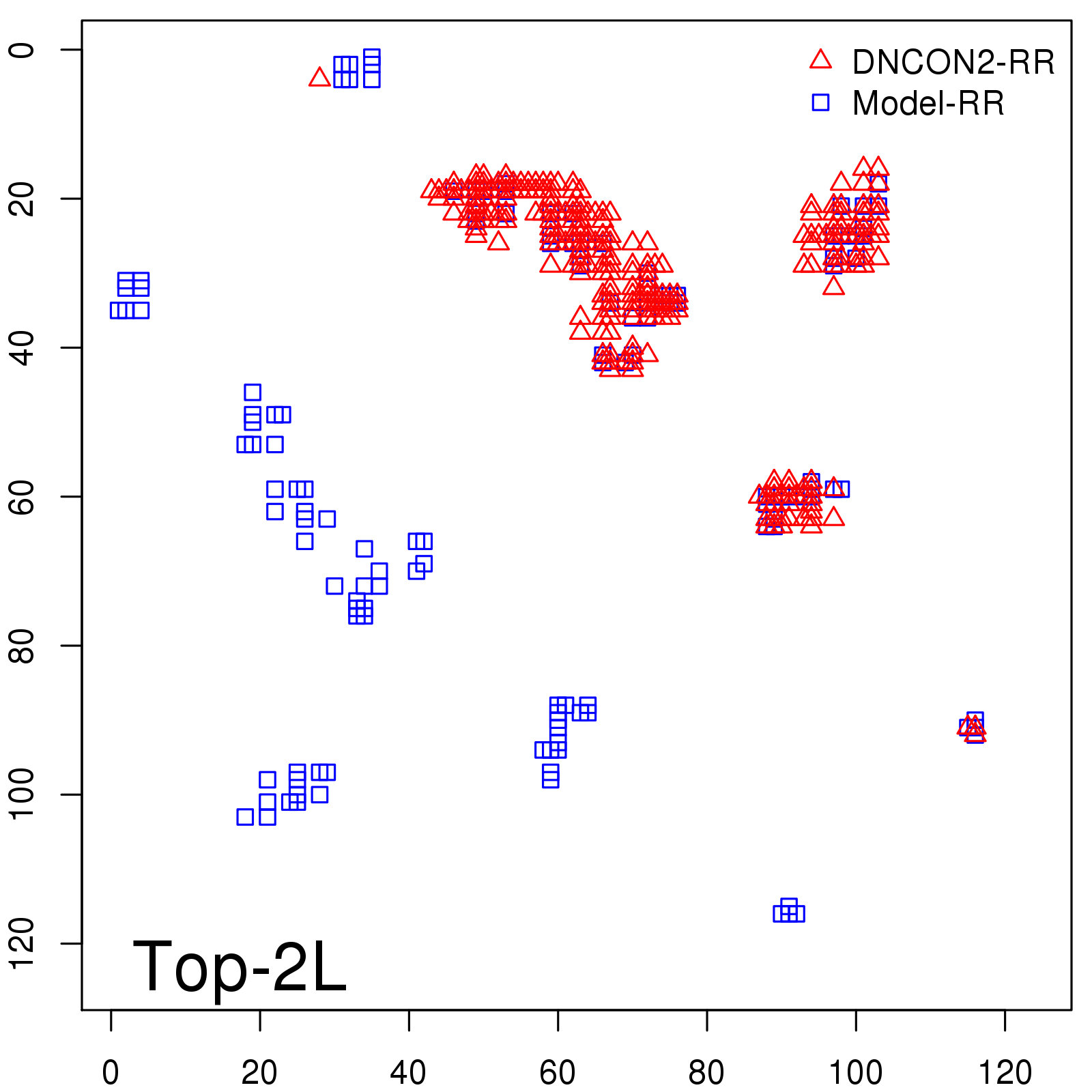
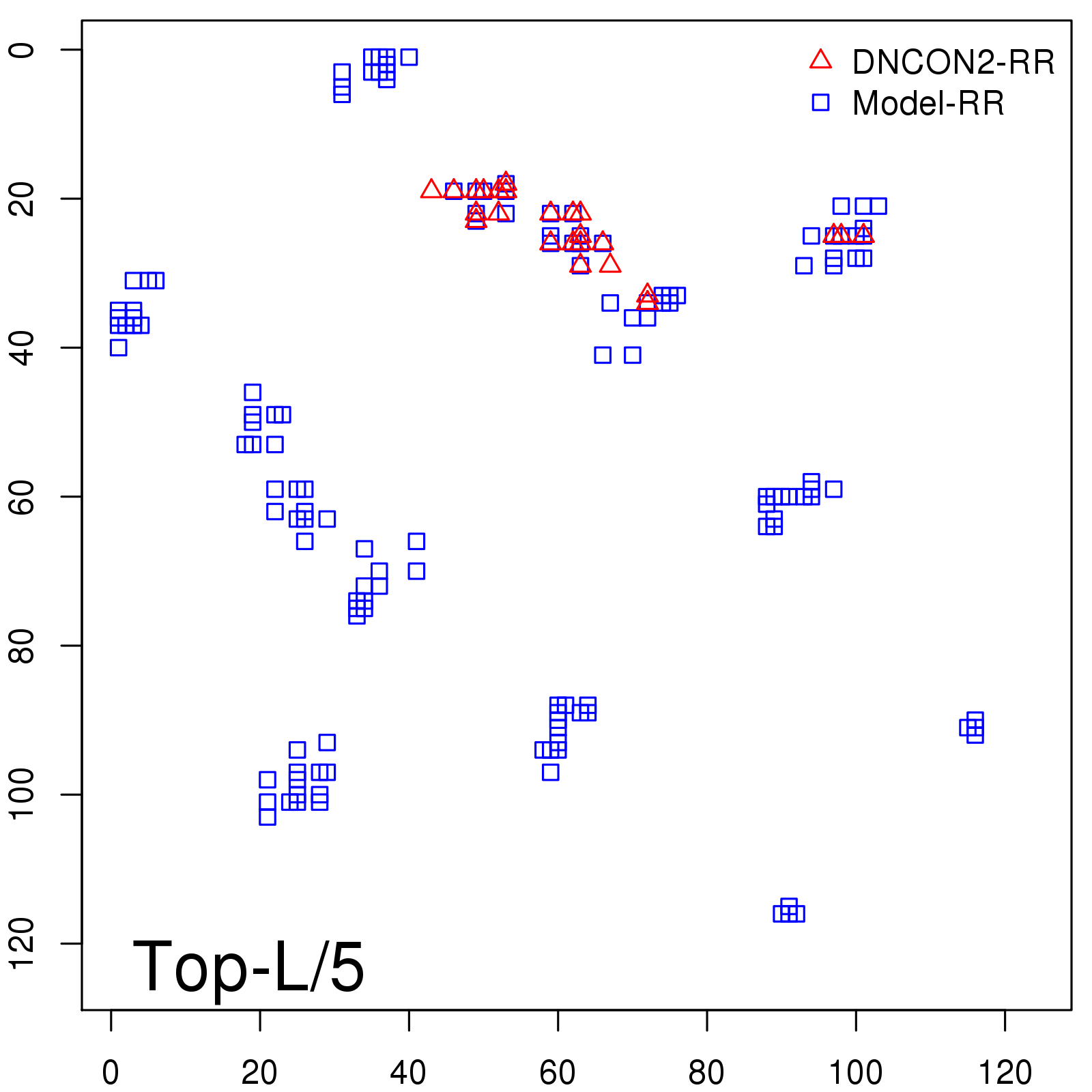
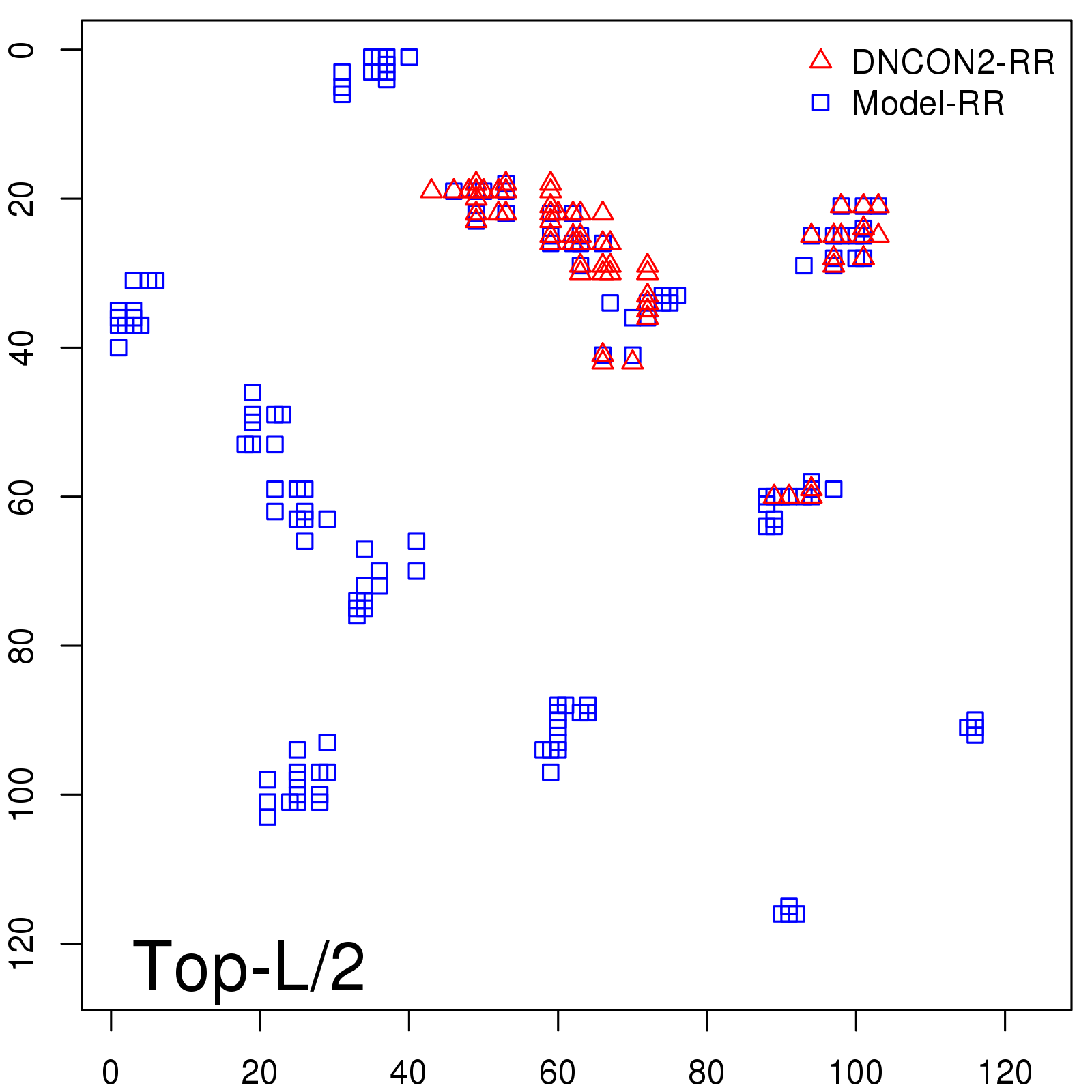
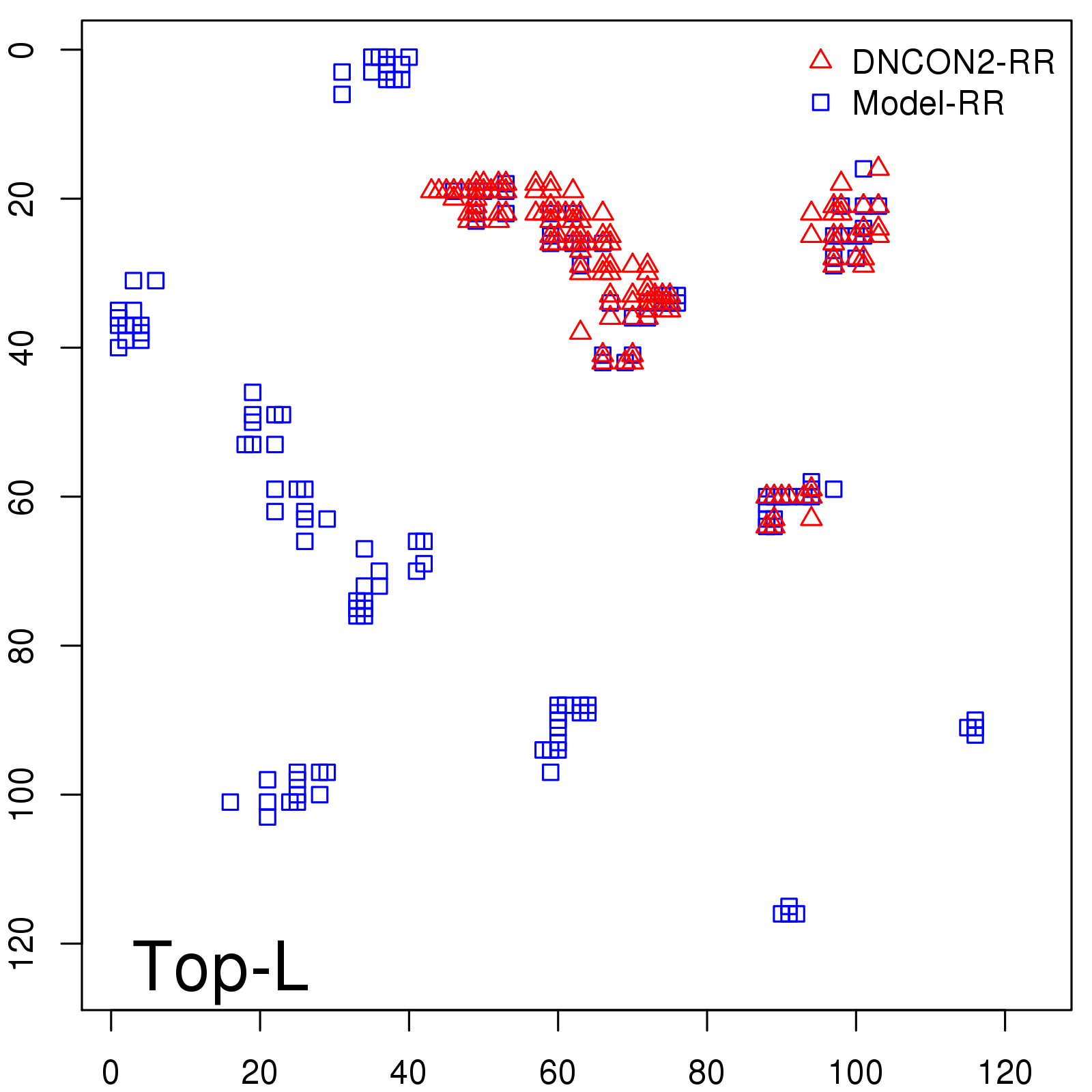
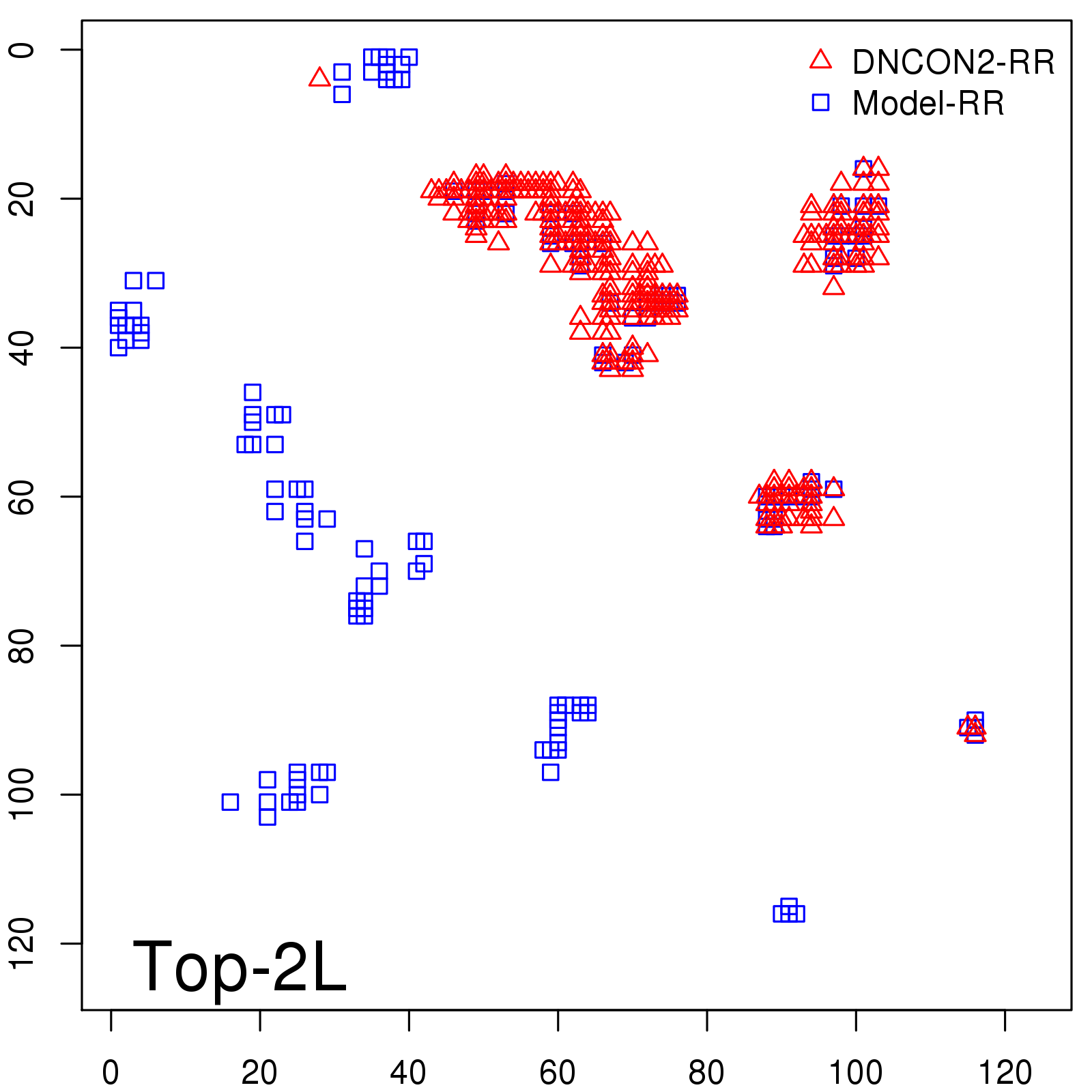
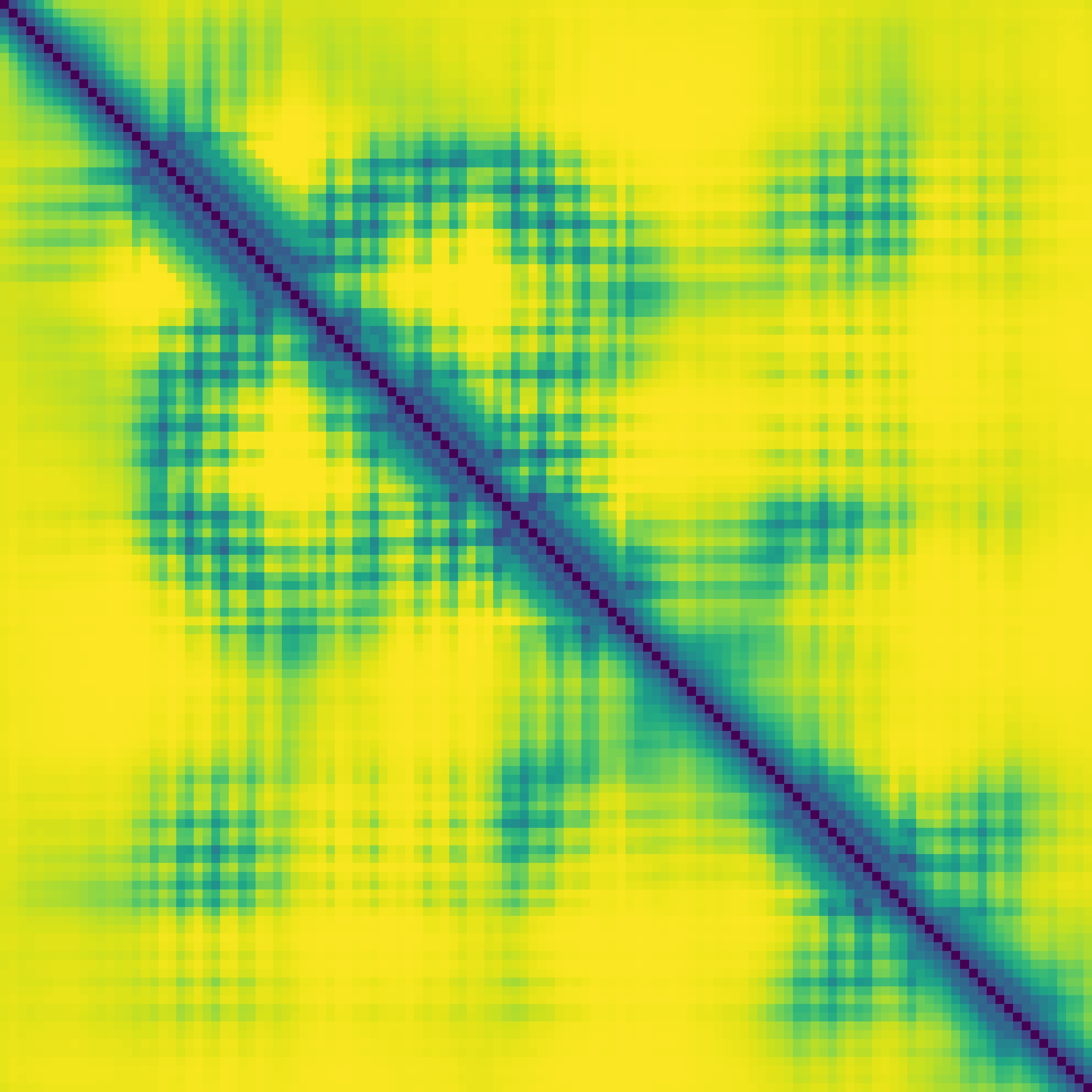
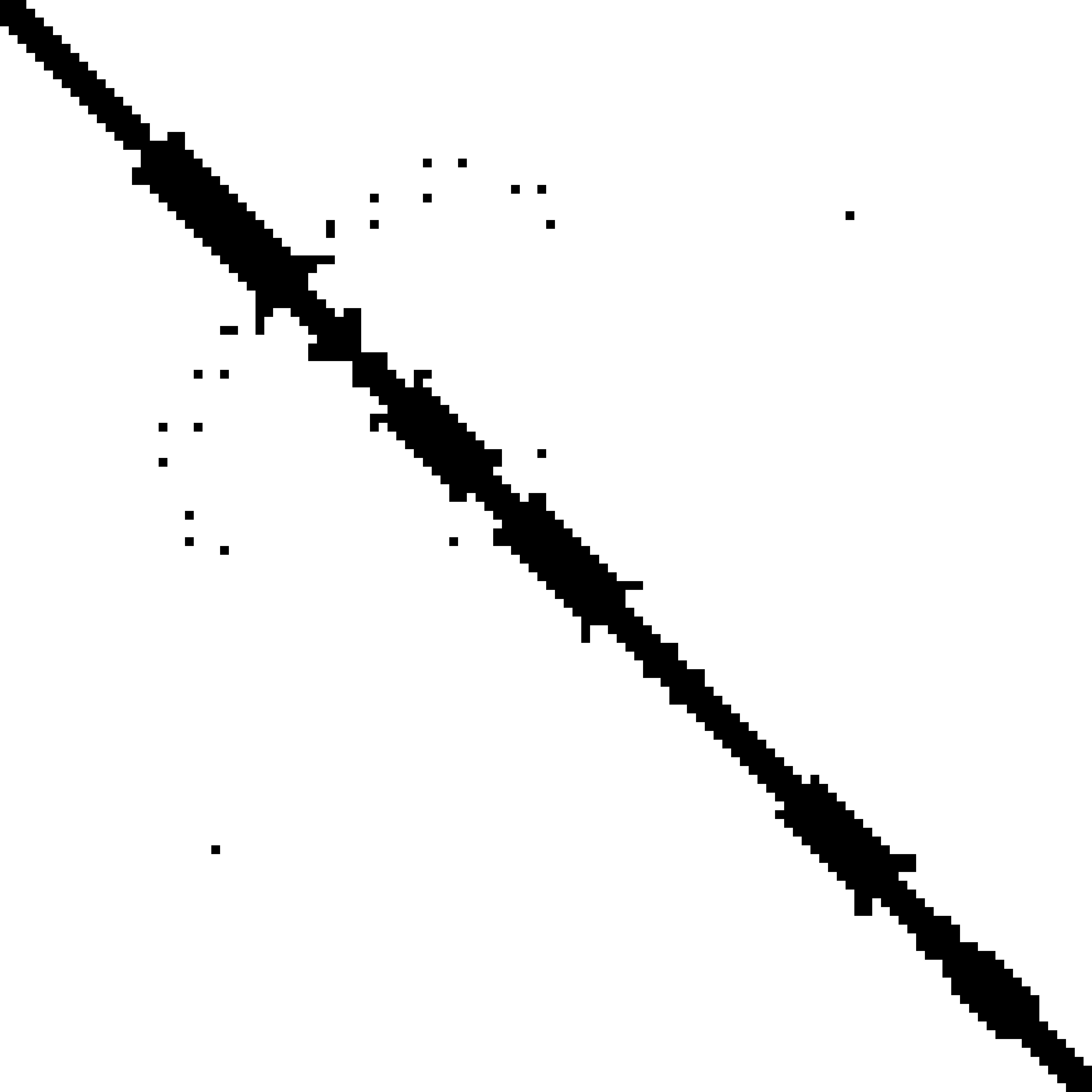
Predicted contact map and distance map
|
Predicted distance map 
|
|
Predicted contact map 
|
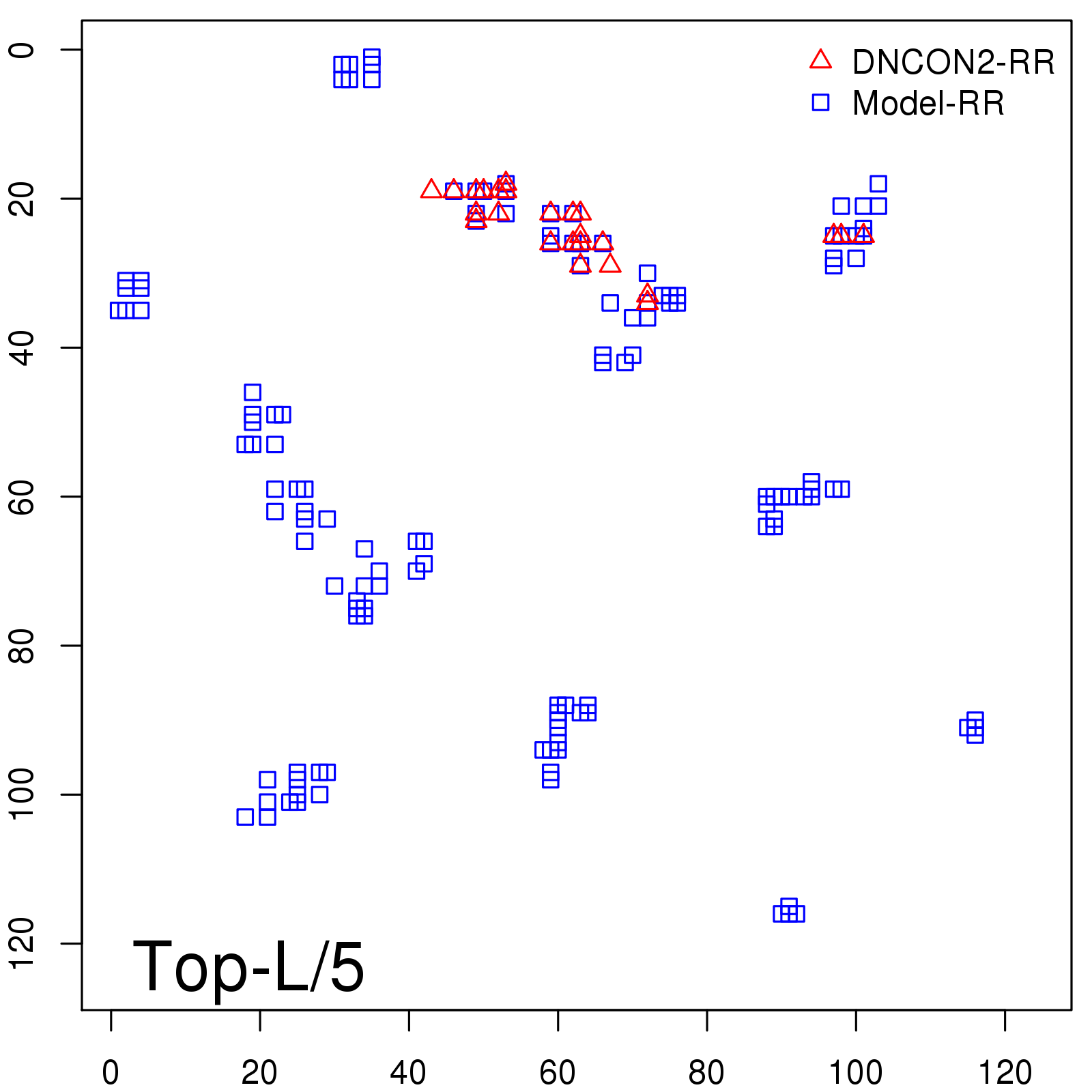
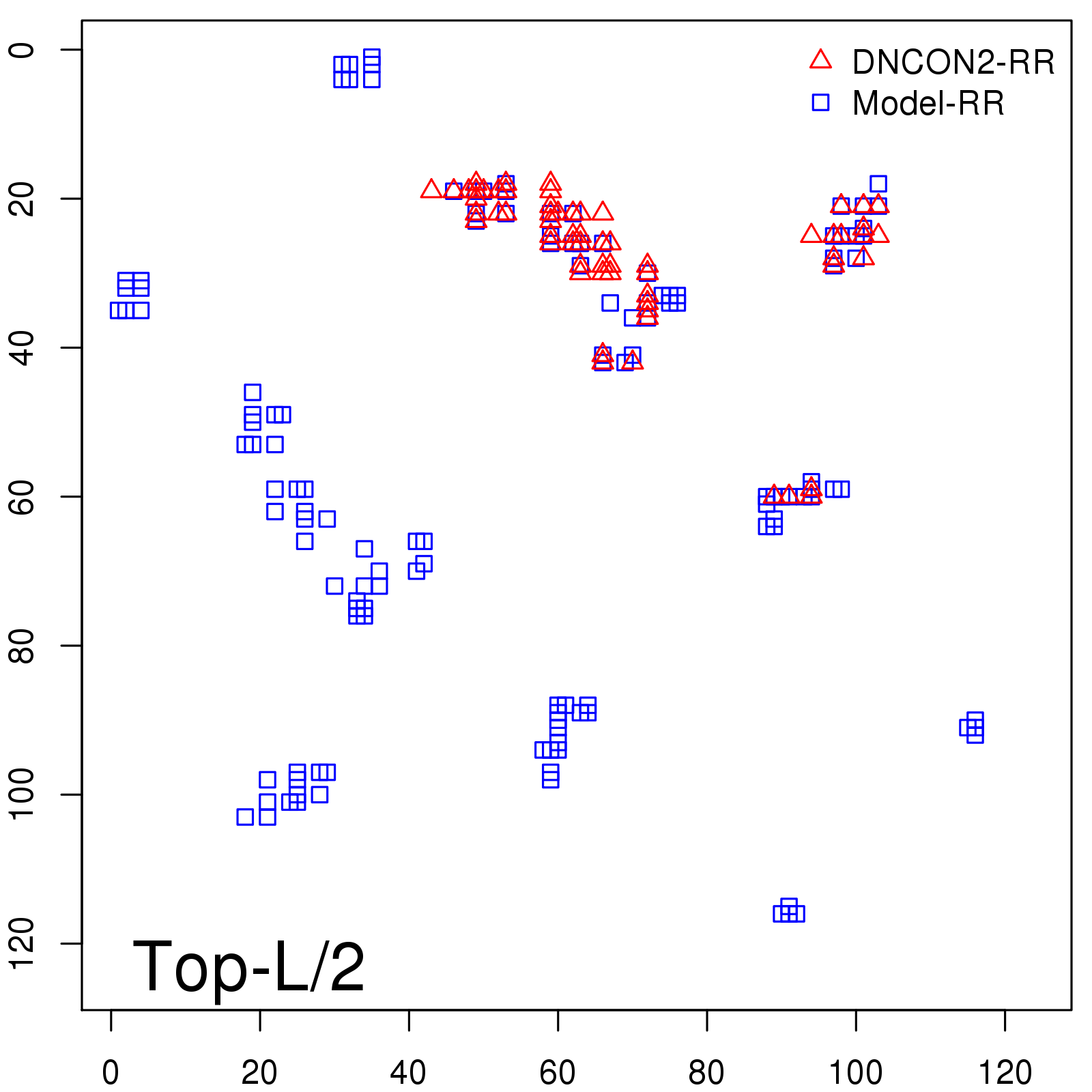
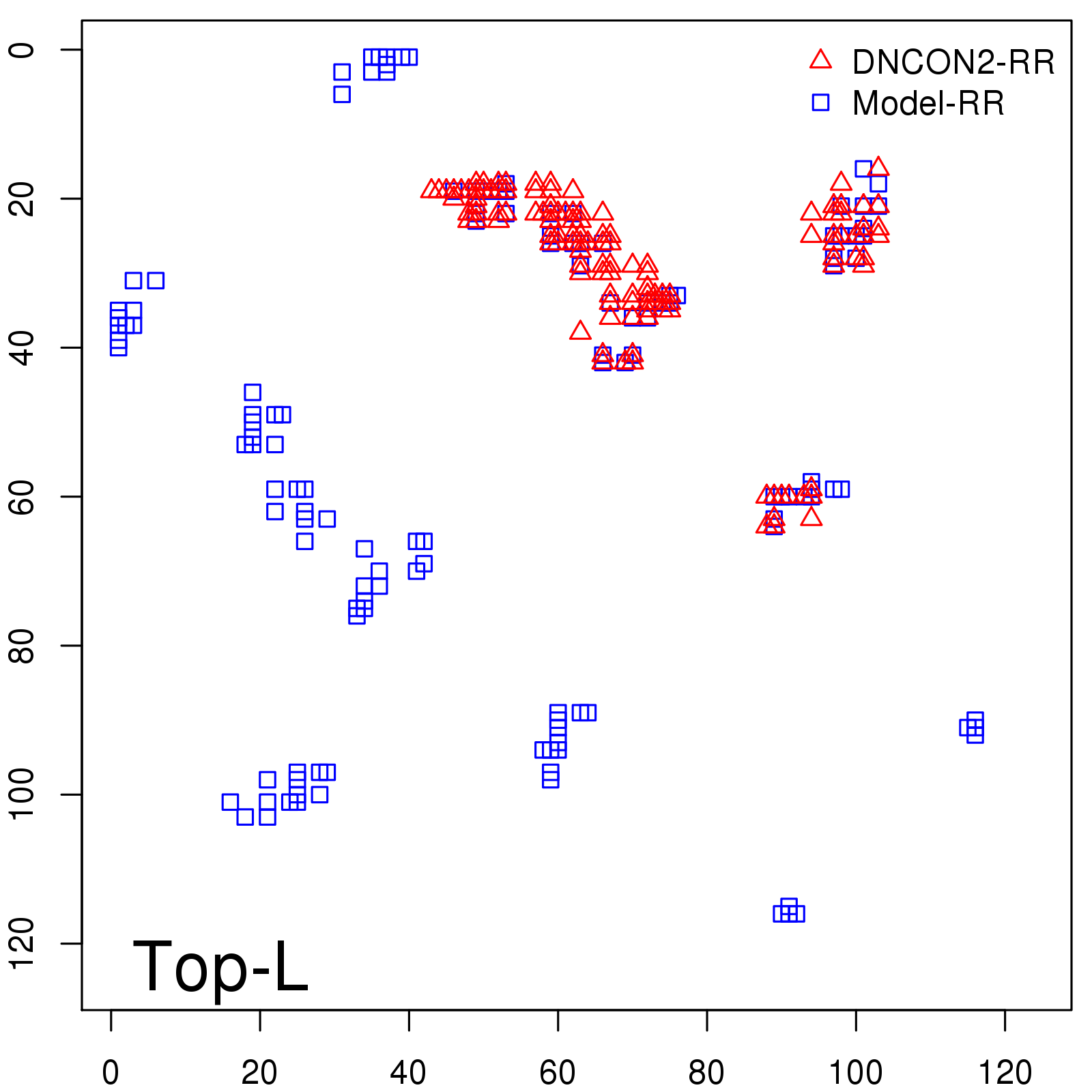
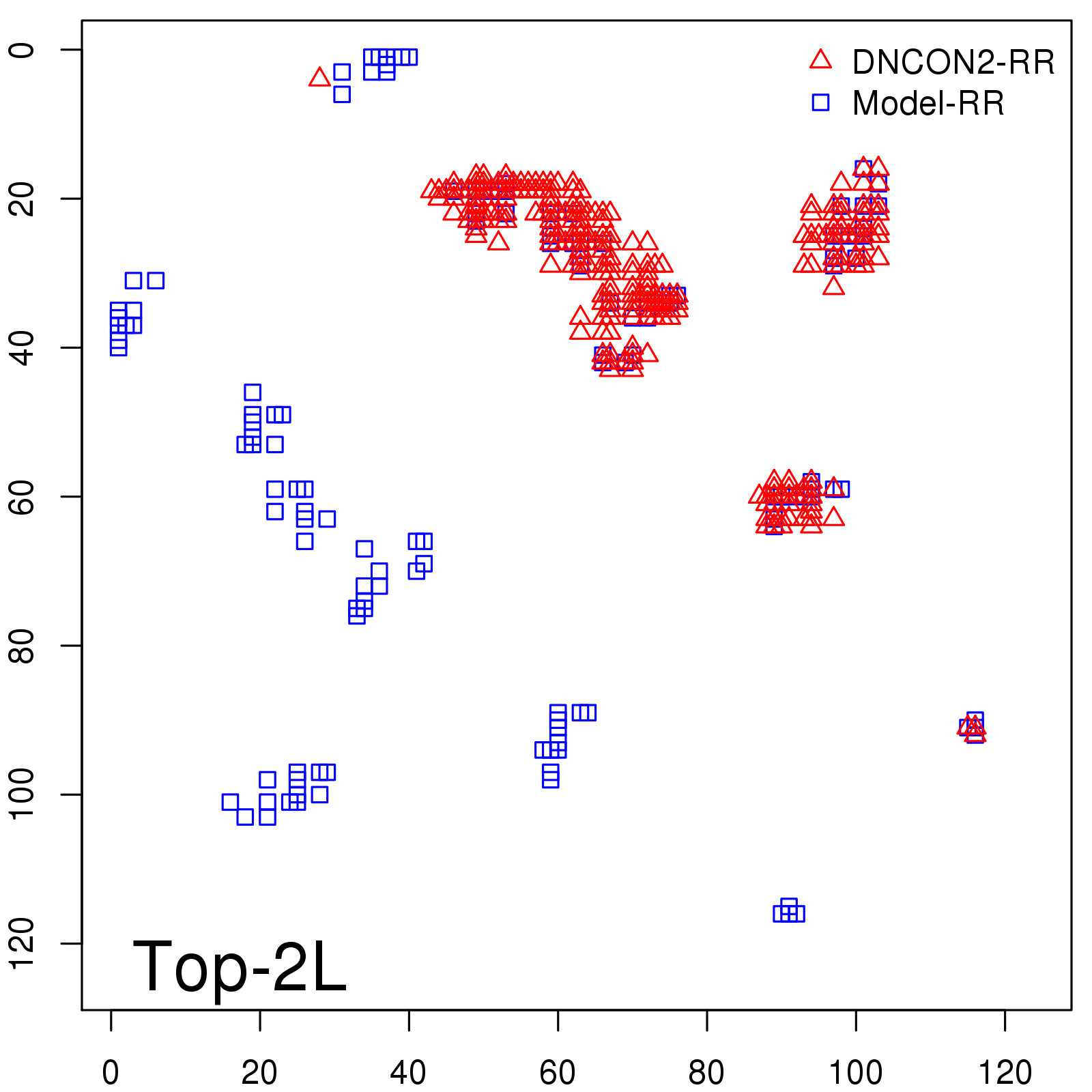
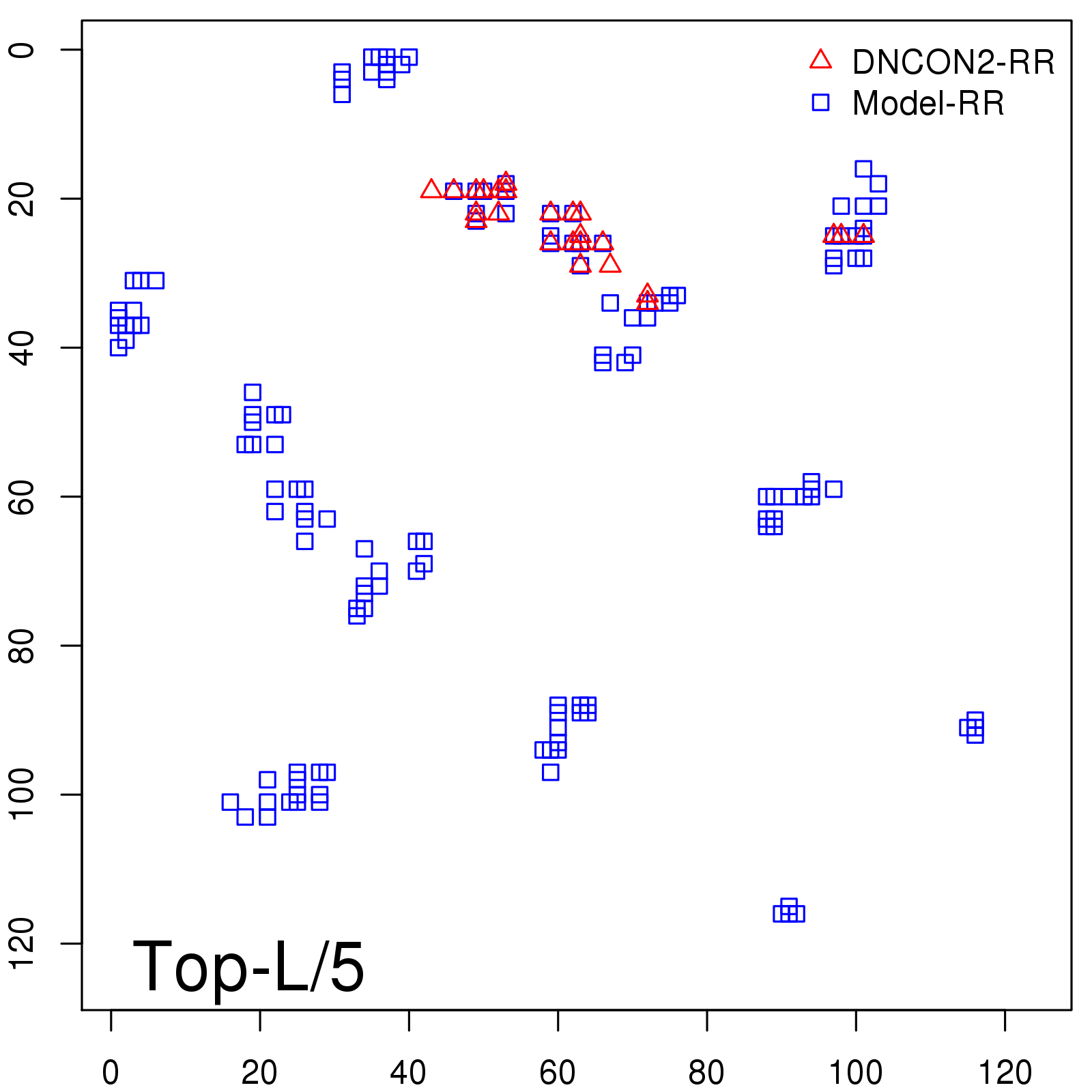
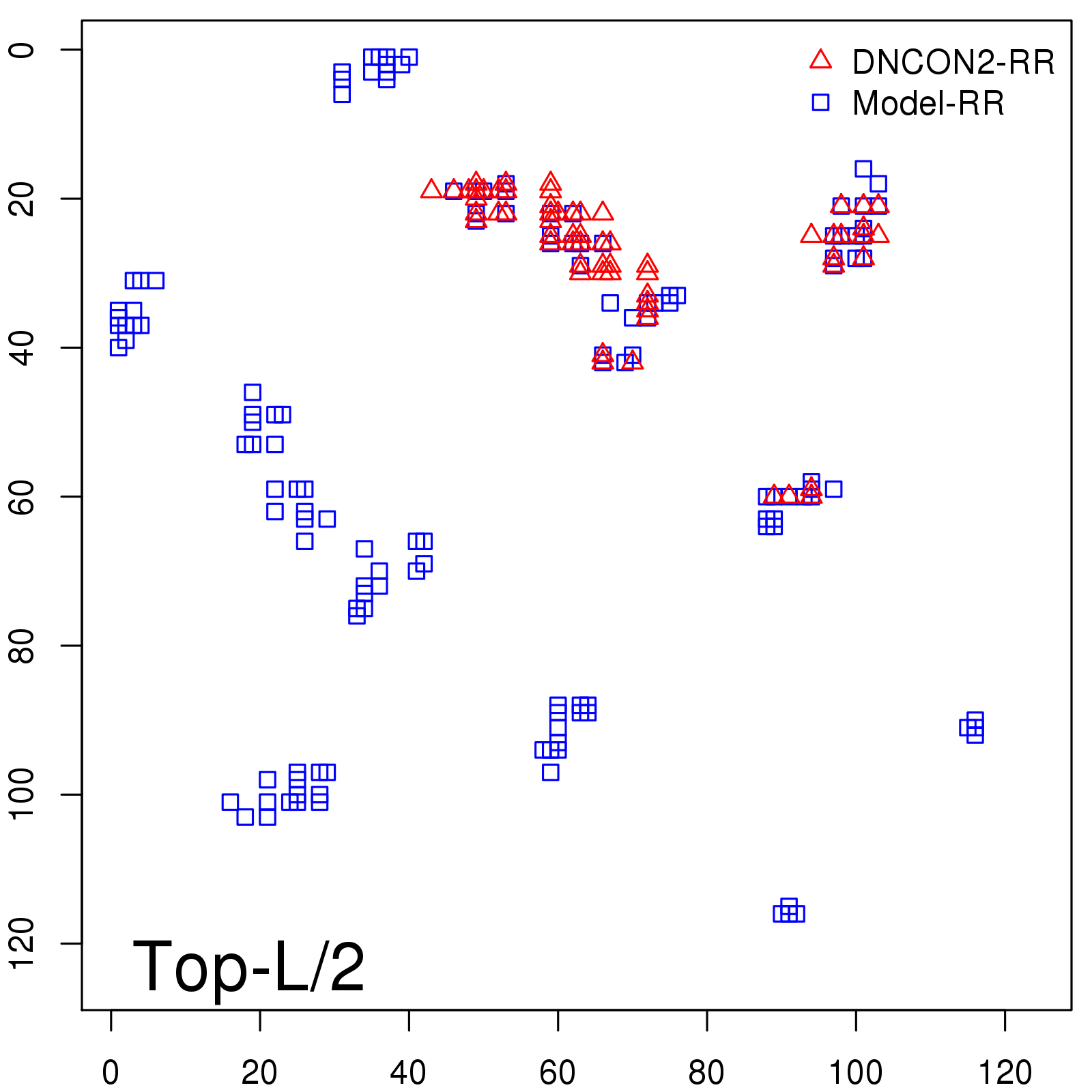
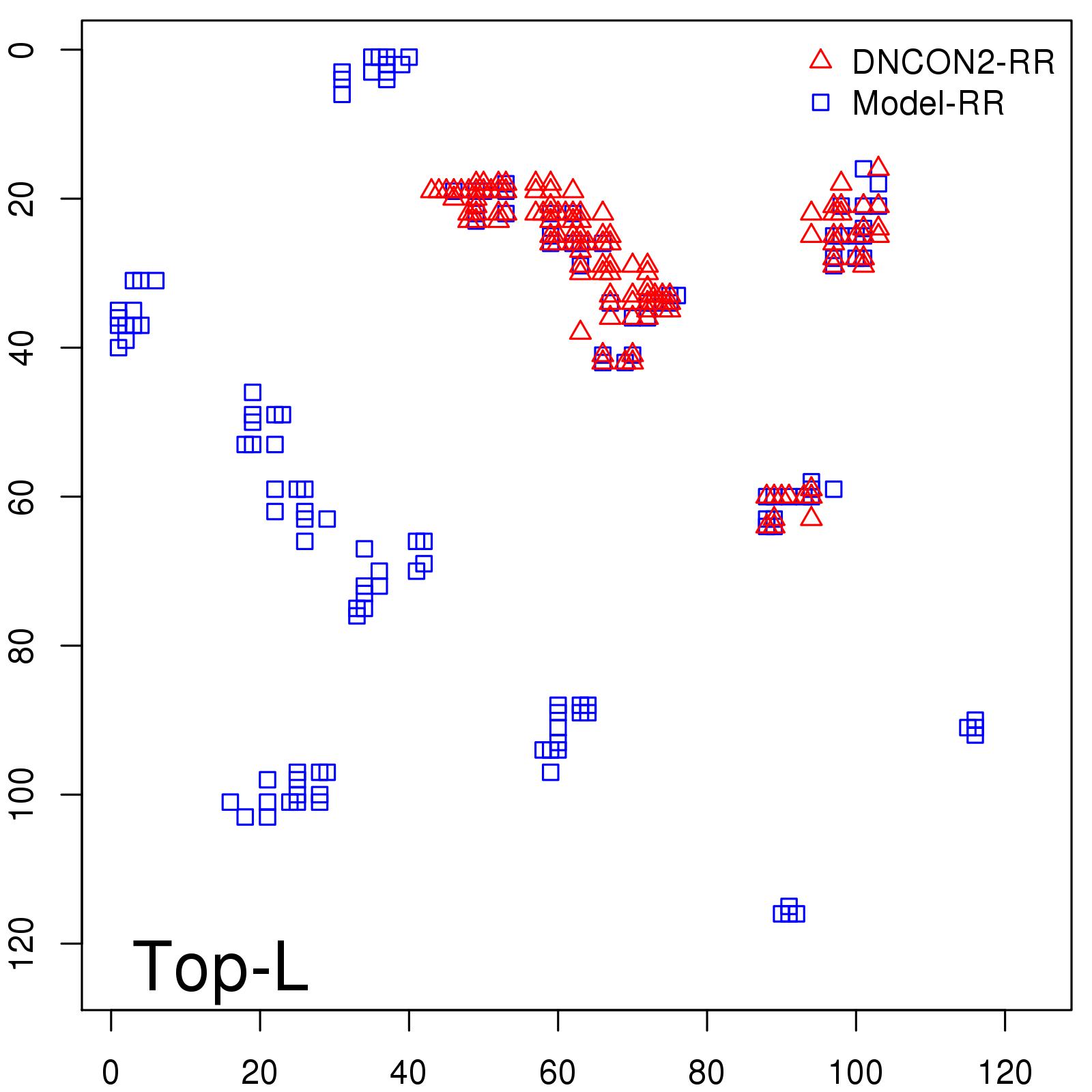
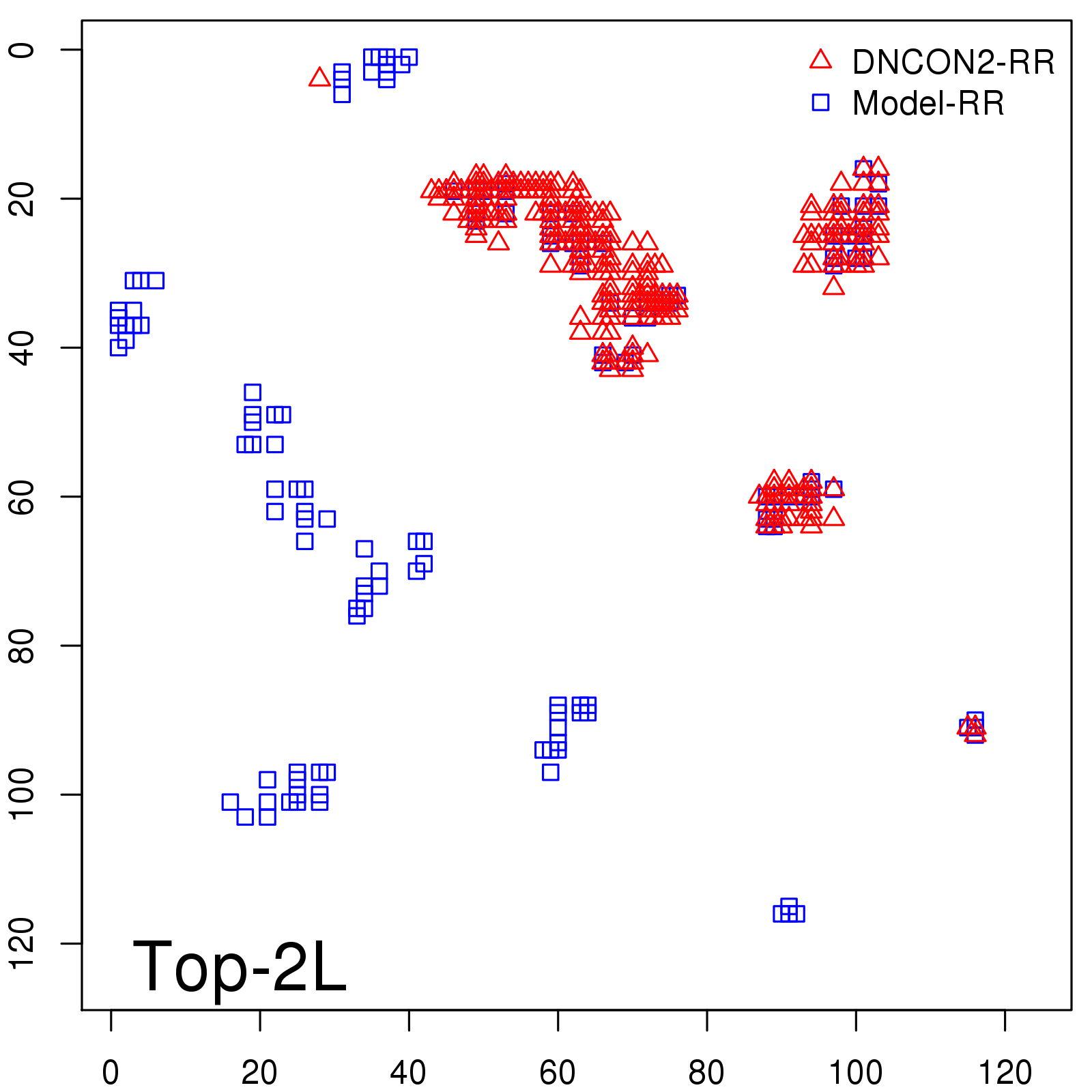
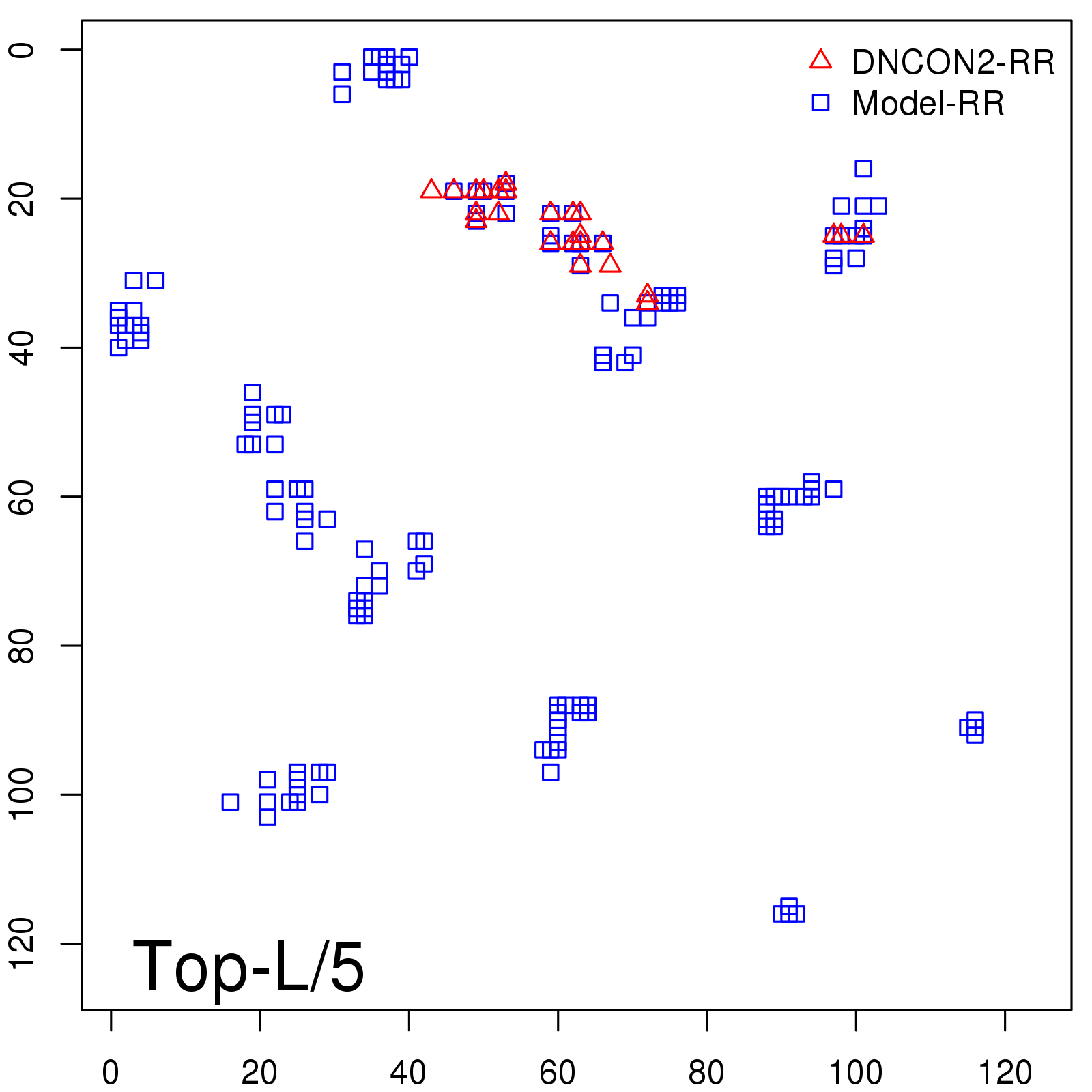
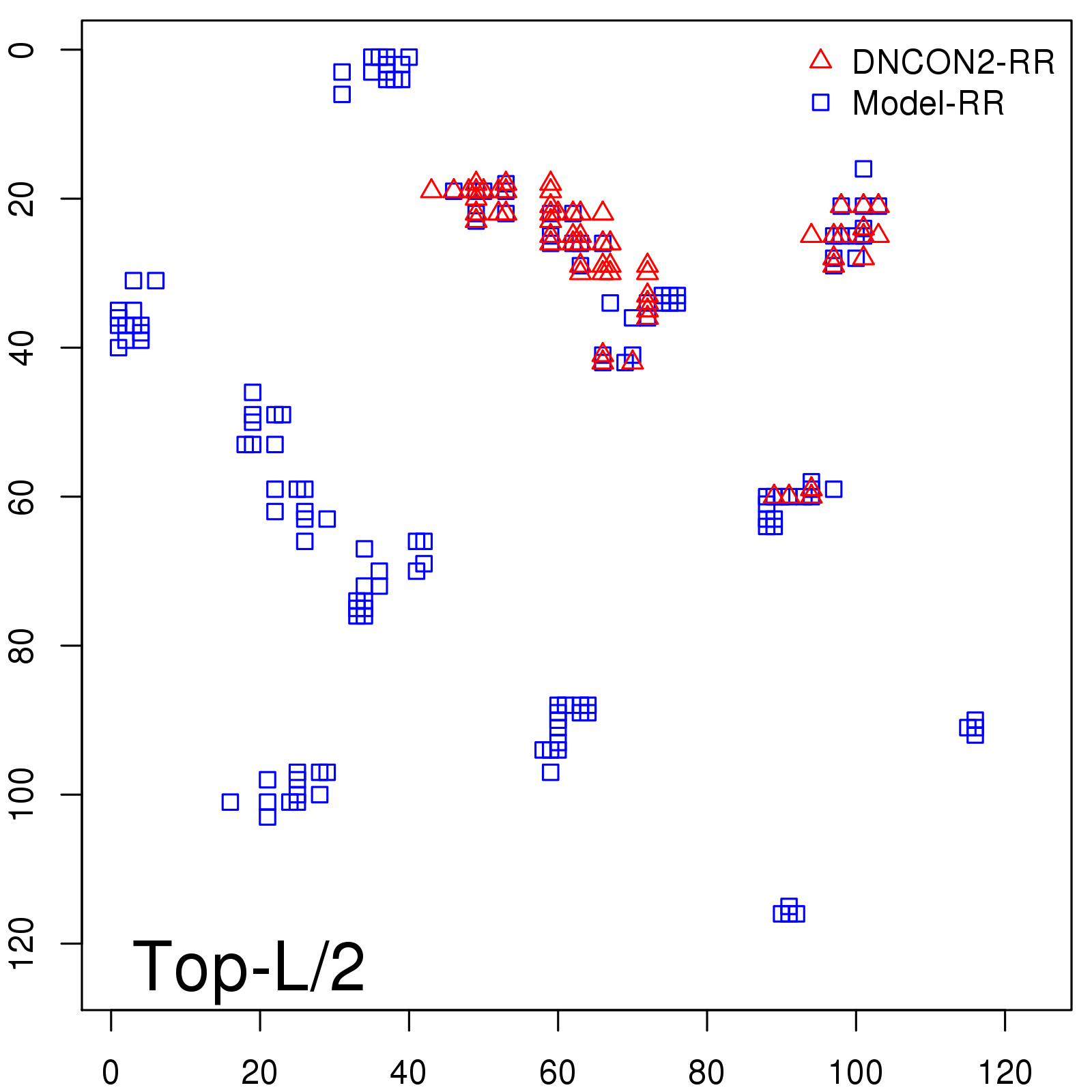
Probability to Precision
| Long-Range |
Average probability |
Predicted precision |
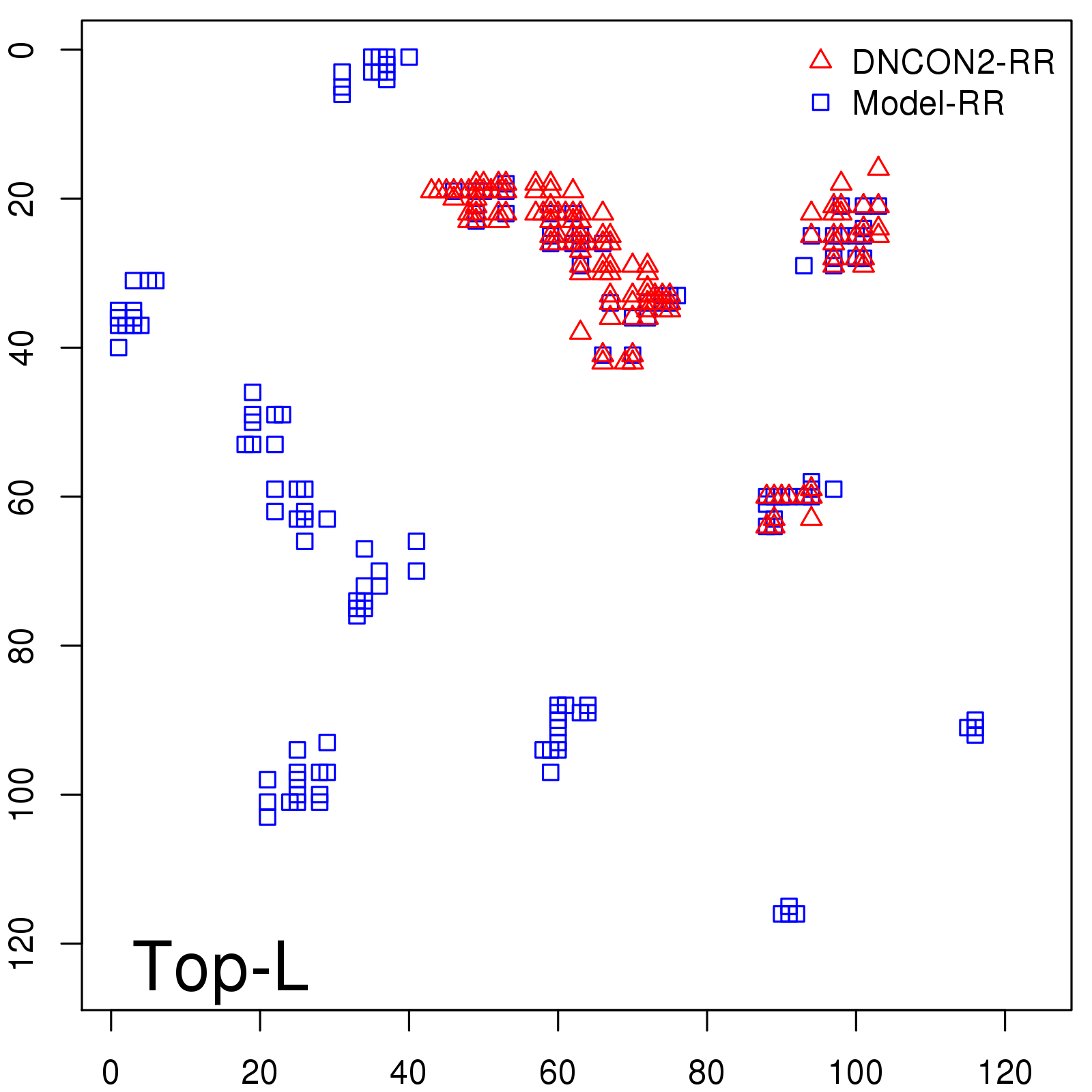
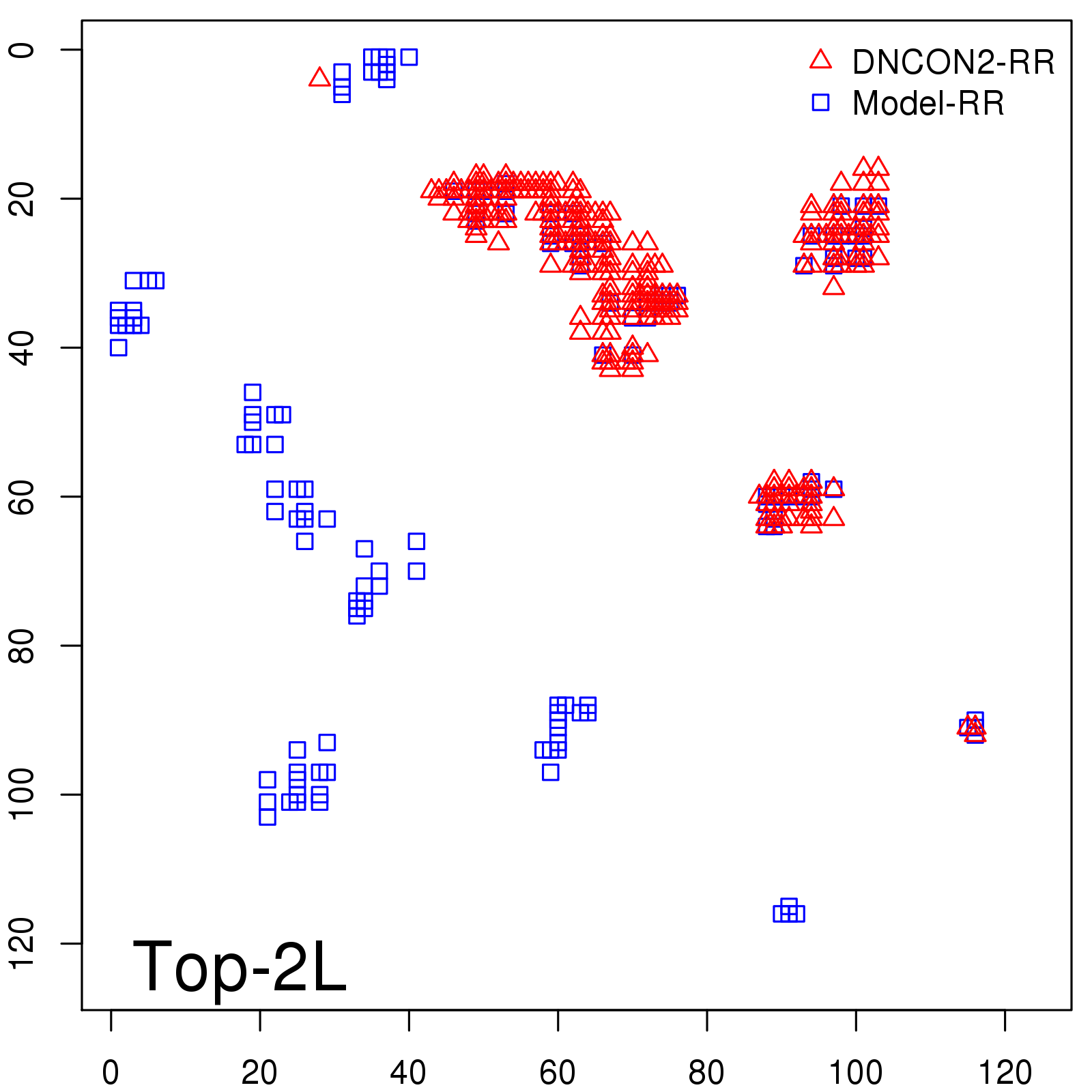
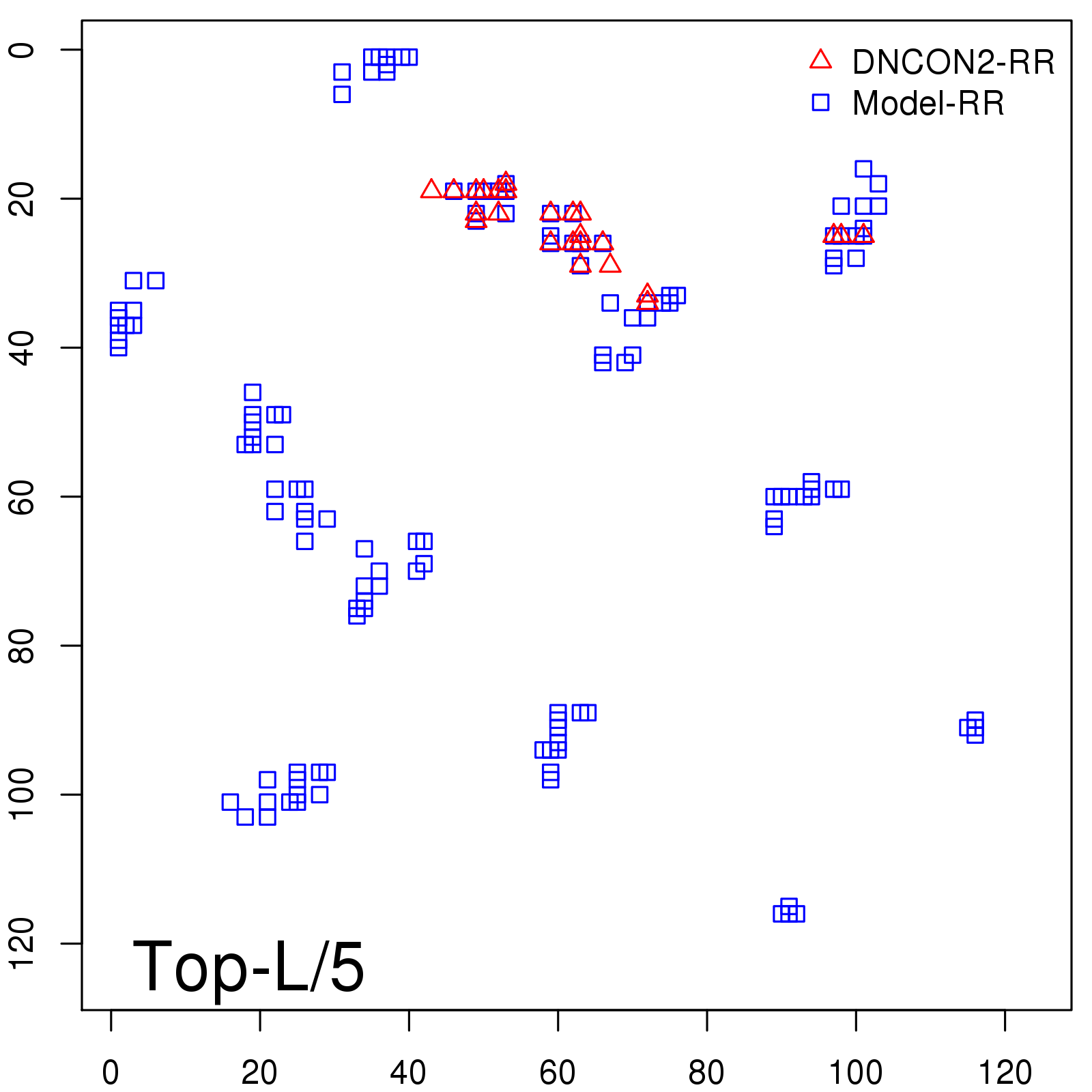
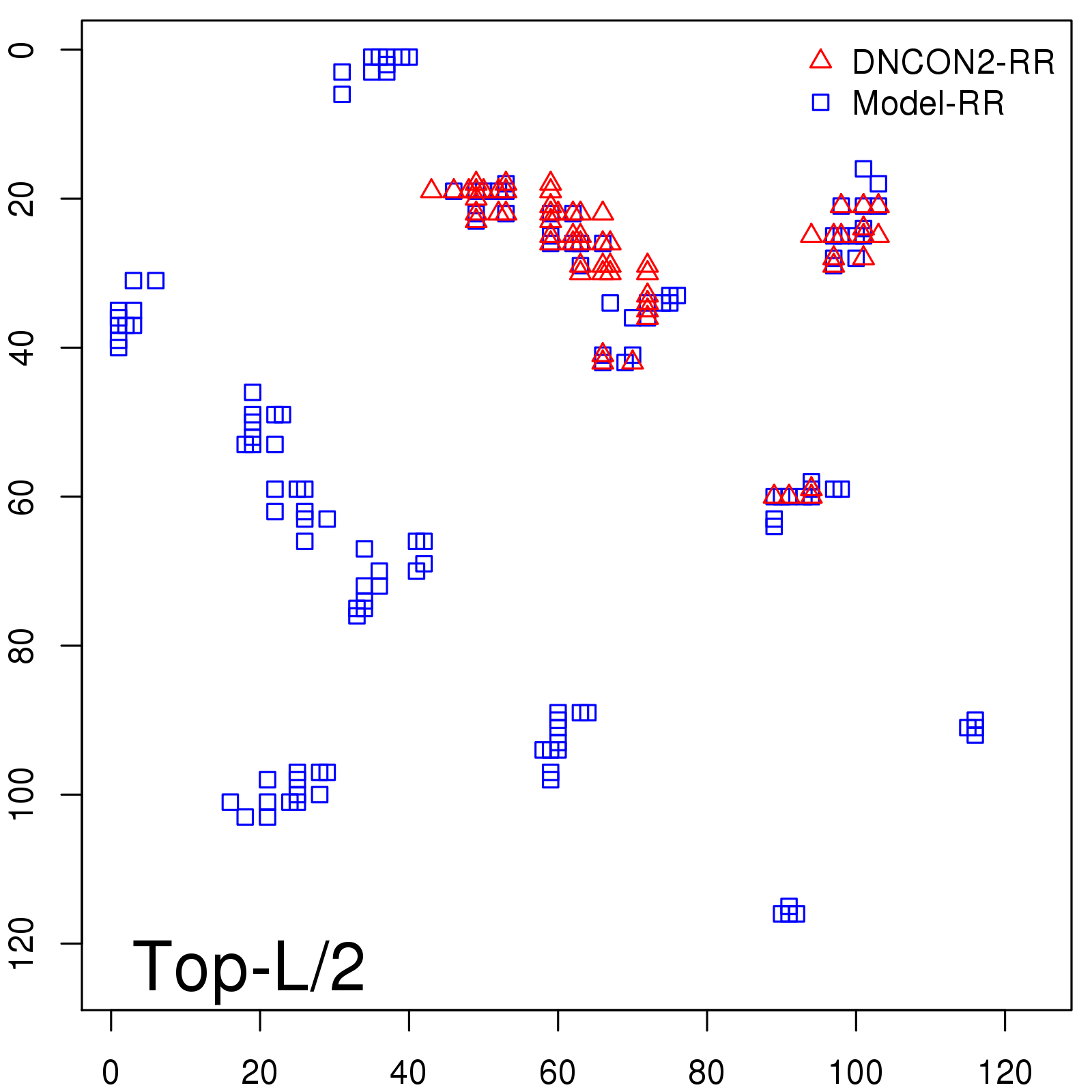
| TopL/5 |
0.46 |
47.71 |
| TopL/2 |
0.34 |
36.85 |
Note: The linear model (Average probability vs Predicted precision) was trained on CASP14 dataset
|
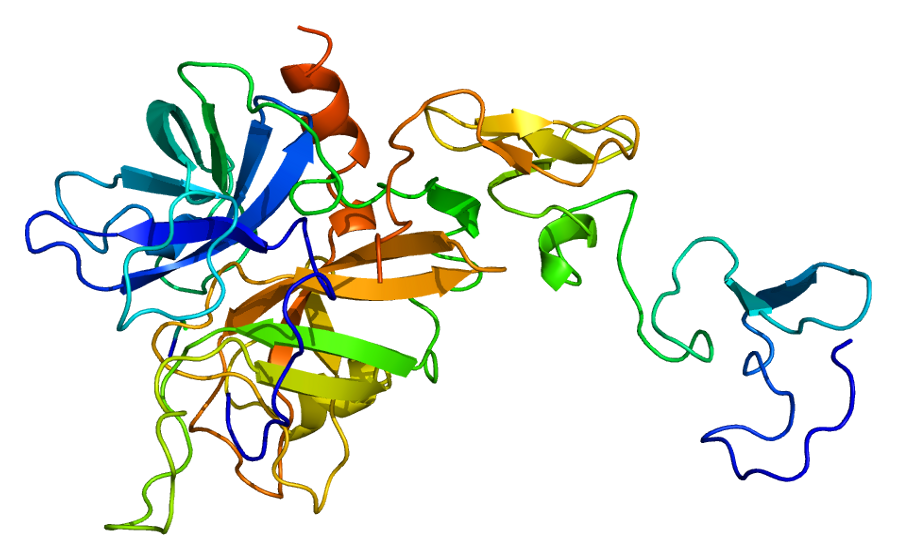
Predicted Top 1 Tertiary structure
|
|
|
| Long-Range |
Precision |
| TopL/5 |
72.00 |
| TopL/2 |
54.84 |
| TopL |
39.52 |
| Top2L |
23.39 |
| Alignment |
Number |
| N |
1086 |
| Neff |
44 |
|
Model comparsion
| Model |
TM-score |
| Model 2 |
0.9054 |
| Model 3 |
0.8977 |
| Model 4 |
0.9181 |
| Model 5 |
0.9225 |
| Average |
0.91092 |
Radius Gyration
|
Similar pdbs
| PDB |
TM-score |
| 3dsi |
0.56648 |
| 3dsj |
0.56596 |
| 2rch |
0.56510 |
| 3dsk |
0.56491 |
| 3cli |
0.56458 |
|
Note: The model is built by ab initio method, no significant templates are found!
|
Predicted Top 2 Tertiary structure
|
|
|
| Long-Range |
Precision |
| TopL/5 |
76.00 |
| TopL/2 |
56.45 |
| TopL |
40.32 |
| Top2L |
23.39 |
| Alignment |
Number |
| N |
1086 |
| Neff |
44 |
|
Model comparsion
| Model |
TM-score |
| Model 1 |
0.9054 |
| Model 3 |
0.9409 |
| Model 4 |
0.9381 |
| Model 5 |
0.9218 |
| Average |
0.92655 |
Radius Gyration
|
Similar pdbs
| PDB |
TM-score |
| 3dsj |
0.55727 |
| 2rcm |
0.55687 |
| 3dsi |
0.55681 |
| 3dsk |
0.55653 |
| 3cli |
0.55524 |
|
Note: The model is built by ab initio method, no significant templates are found!
|
Predicted Top 3 Tertiary structure
|
|
|
| Long-Range |
Precision |
| TopL/5 |
76.00 |
| TopL/2 |
54.84 |
| TopL |
37.90 |
| Top2L |
22.18 |
| Alignment |
Number |
| N |
1086 |
| Neff |
44 |
|
Model comparsion
| Model |
TM-score |
| Model 1 |
0.8977 |
| Model 2 |
0.9409 |
| Model 4 |
0.9269 |
| Model 5 |
0.9188 |
| Average |
0.92107 |
Radius Gyration
|
Similar pdbs
| PDB |
TM-score |
| 2o36 |
0.55800 |
| 2rcm |
0.55740 |
| 3dsi |
0.55706 |
| 1s4b |
0.55696 |
| 3dsk |
0.55685 |
|
Note: The model is built by ab initio method, no significant templates are found!
|
Predicted Top 4 Tertiary structure
|
|
|
| Long-Range |
Precision |
| TopL/5 |
72.00 |
| TopL/2 |
54.84 |
| TopL |
38.71 |
| Top2L |
22.98 |
| Alignment |
Number |
| N |
1086 |
| Neff |
44 |
|
Model comparsion
| Model |
TM-score |
| Model 1 |
0.9181 |
| Model 2 |
0.9381 |
| Model 3 |
0.9269 |
| Model 5 |
0.9194 |
| Average |
0.92563 |
Radius Gyration
|
Similar pdbs
| PDB |
TM-score |
| 3dsi |
0.55581 |
| 3dsj |
0.55518 |
| 3cli |
0.55459 |
| 2rch |
0.55435 |
| 3dsk |
0.55415 |
|
Note: The model is built by ab initio method, no significant templates are found!
|
Predicted Top 5 Tertiary structure
|
|
|
| Long-Range |
Precision |
| TopL/5 |
72.00 |
| TopL/2 |
53.23 |
| TopL |
39.52 |
| Top2L |
23.79 |
| Alignment |
Number |
| N |
1086 |
| Neff |
44 |
|
Model comparsion
| Model |
TM-score |
| Model 1 |
0.9225 |
| Model 2 |
0.9218 |
| Model 3 |
0.9188 |
| Model 4 |
0.9194 |
| Average |
0.92063 |
Radius Gyration
|
Similar pdbs
| PDB |
TM-score |
| 3dsi |
0.55608 |
| 3cli |
0.55607 |
| 3dsj |
0.55588 |
| 3dsk |
0.55526 |
| 2rch |
0.55490 |
|
Note: The model is built by ab initio method, no significant templates are found!
|
References
[1] Hou, J., Wu, T., Cao, R., & Cheng, J. (2019). Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. bioRxiv, 552422.
[2] Li, J., Deng, X., Eickholt, J., & Cheng, J. (2013). Designing and benchmarking the MULTICOM protein structure prediction system. BMC structural biology, 13(1), 2.
[3] Cheng, J., Li, J., Wang, Z., Eickholt, J., & Deng, X. (2012). The MULTICOM toolbox for protein structure prediction. BMC bioinformatics, 13(1), 65.
[4] Wang, Z., Eickholt, J., & Cheng, J. (2010). MULTICOM: a multi-level combination approach to protein structure prediction and its assessments in CASP8. Bioinformatics, 26(7), 882-888.
 )
) )
)
 )
) )
) )
) )
) )
)