Results of Structure Prediction for Target Name: P76148 ( Click  )
)
Domain Boundary prediction ( View  )
)

Protein sequence
>P76148: 1-119| 1-60: | M | Q | S | L | D | P | L | F | A | R | L | S | R | S | K | F | R | S | R | F | R | L | G | M | K | E | R | Q | Y | C | L | E | K | G | A | P | V | I | E | Q | H | A | A | D | F | V | A | K | R | L | A | P | A | L | P | A | N | D | G | K |
| 61-119: | Q | T | P | M | R | G | H | P | V | F | I | A | Q | H | A | T | A | T | C | C | R | G | C | L | A | K | W | H | N | I | P | Q | G | V | S | L | S | E | E | Q | Q | R | Y | I | V | A | V | I | Y | H | W | L | V | V | Q | M | N | Q | P |
Secondary structure prediction (H: Helix E: Strand C: Coil)
| 1-60: | C | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | E | E | C | C | C | H | H | H | H | H | H | H | H | H | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | C | C |
| 61-119: | C | C | C | C | C | C | C | C | E | E | E | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | |
| H(Helix): 74(62.18%) | E(Strand): 5(4.2%) | C(Coil): 40(33.61%) |
Solvent accessibility prediction (e: Exposed b: Buried)
| 1-60: | E | E | E | B | E | E | B | B | E | E | B | E | E | E | E | B | B | B | E | B | E | B | E | E | E | B | B | E | B | B | E | E | E | E | B | E | E | B | E | E | B | B | E | E | B | B | E | E | E | B | E | E | E | E | B | E | E | E | B | E |
| 61-119: | E | B | B | E | E | E | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | E | B | B | E | E | B | B | E | B | E | E | E | E | E | B | E | E | E | B | B | E | B | B | B | E | B | B | B | E | B | B | E | E | E | B | E | E | E | |
| e(Exposed): 63(52.94%) | b(Buried): 56(47.06%) |
Disorder prediction (N: Normal T: Disorder)
| 1-60: | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 61-119: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | |
| N(Normal): 118(99.16%) | T(Disorder): 1(0.84%) |
Select domain
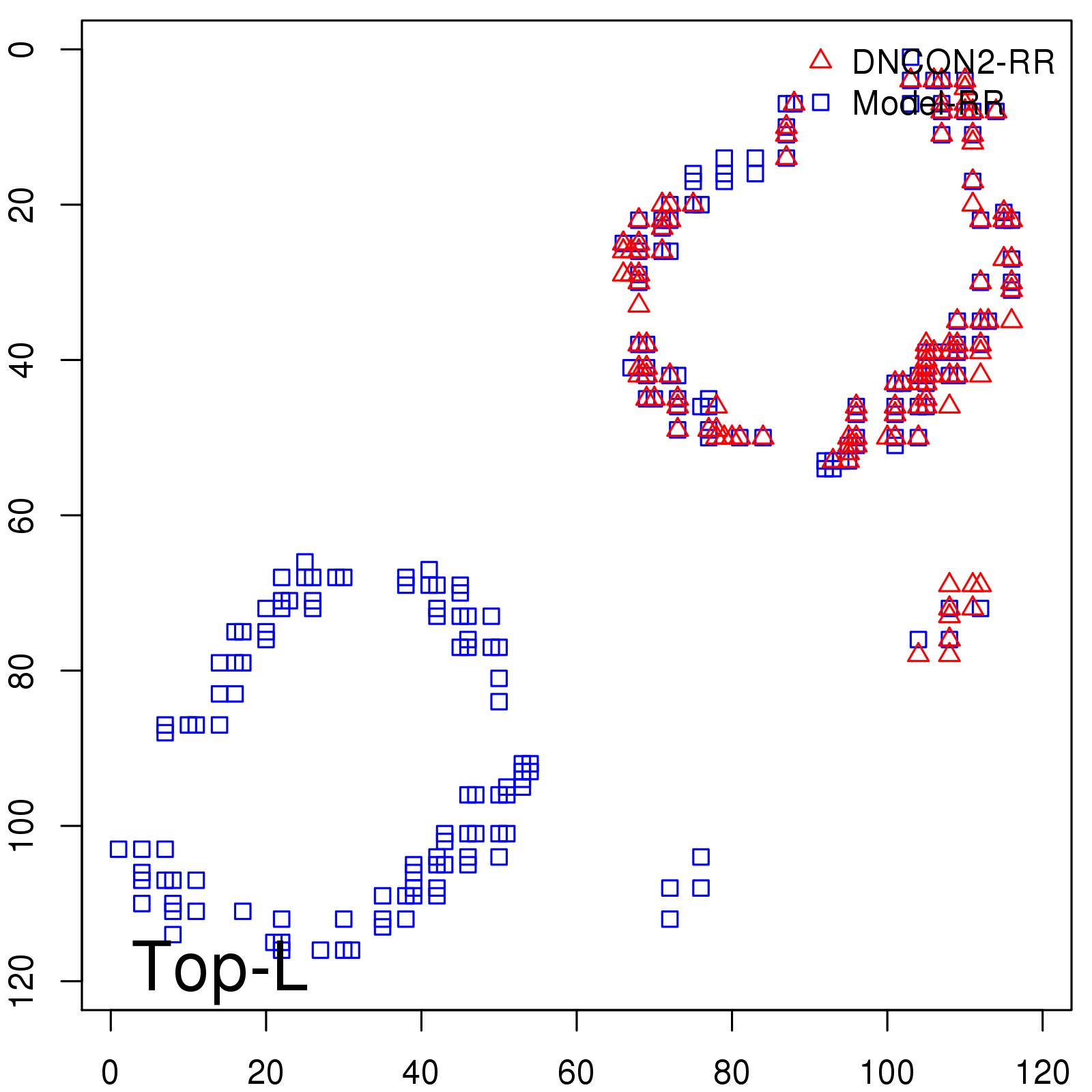
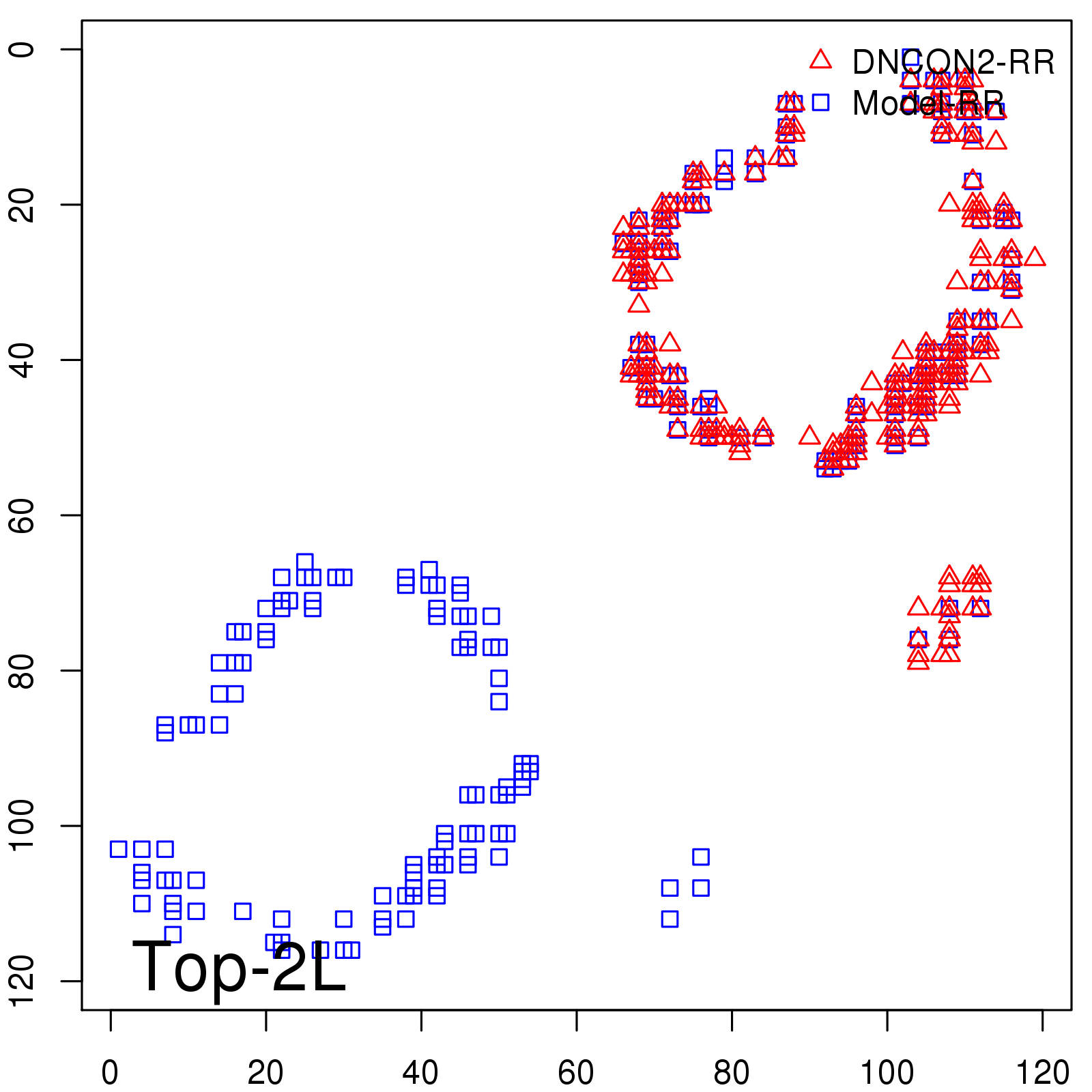
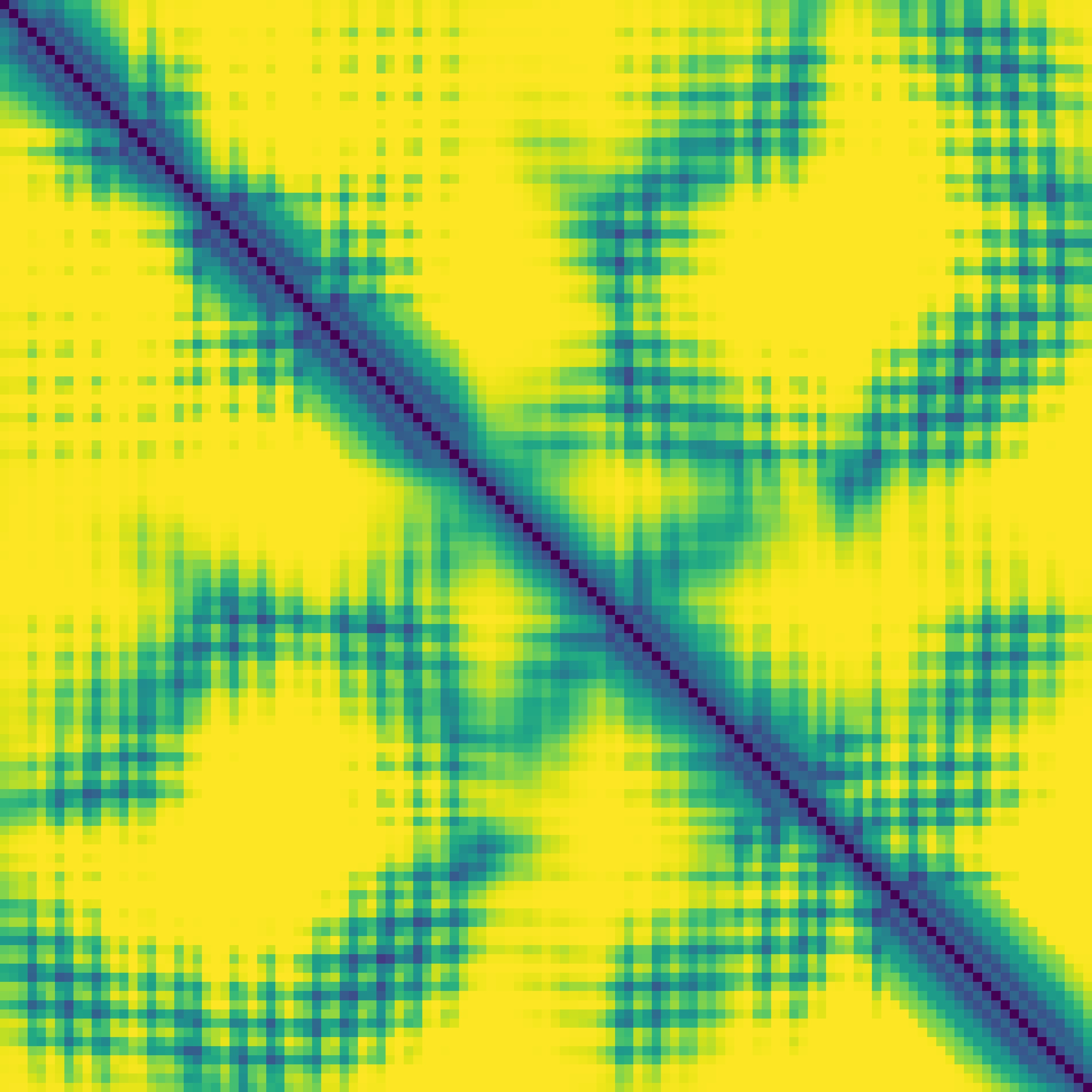
Predicted contact map and distance map
|
|
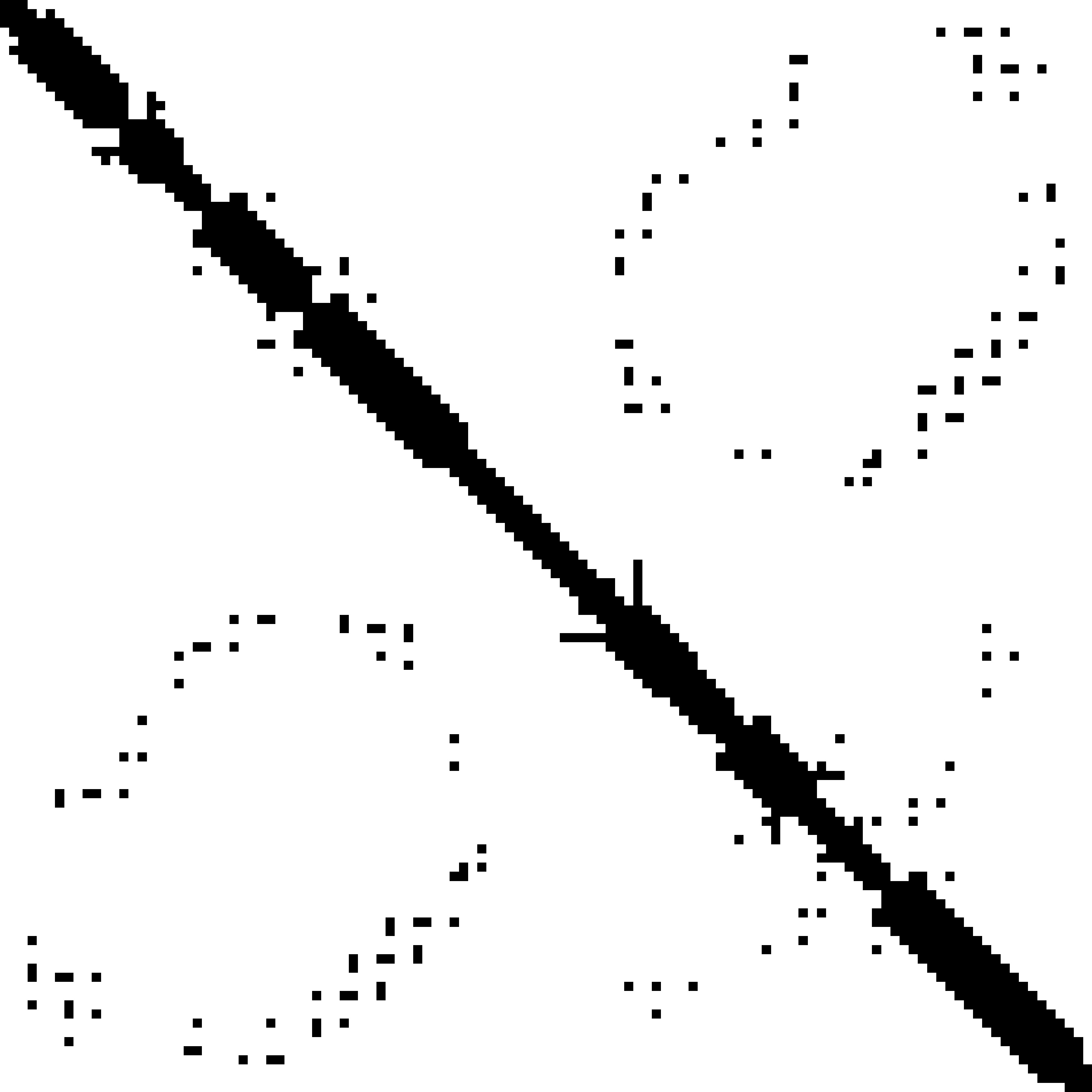
|
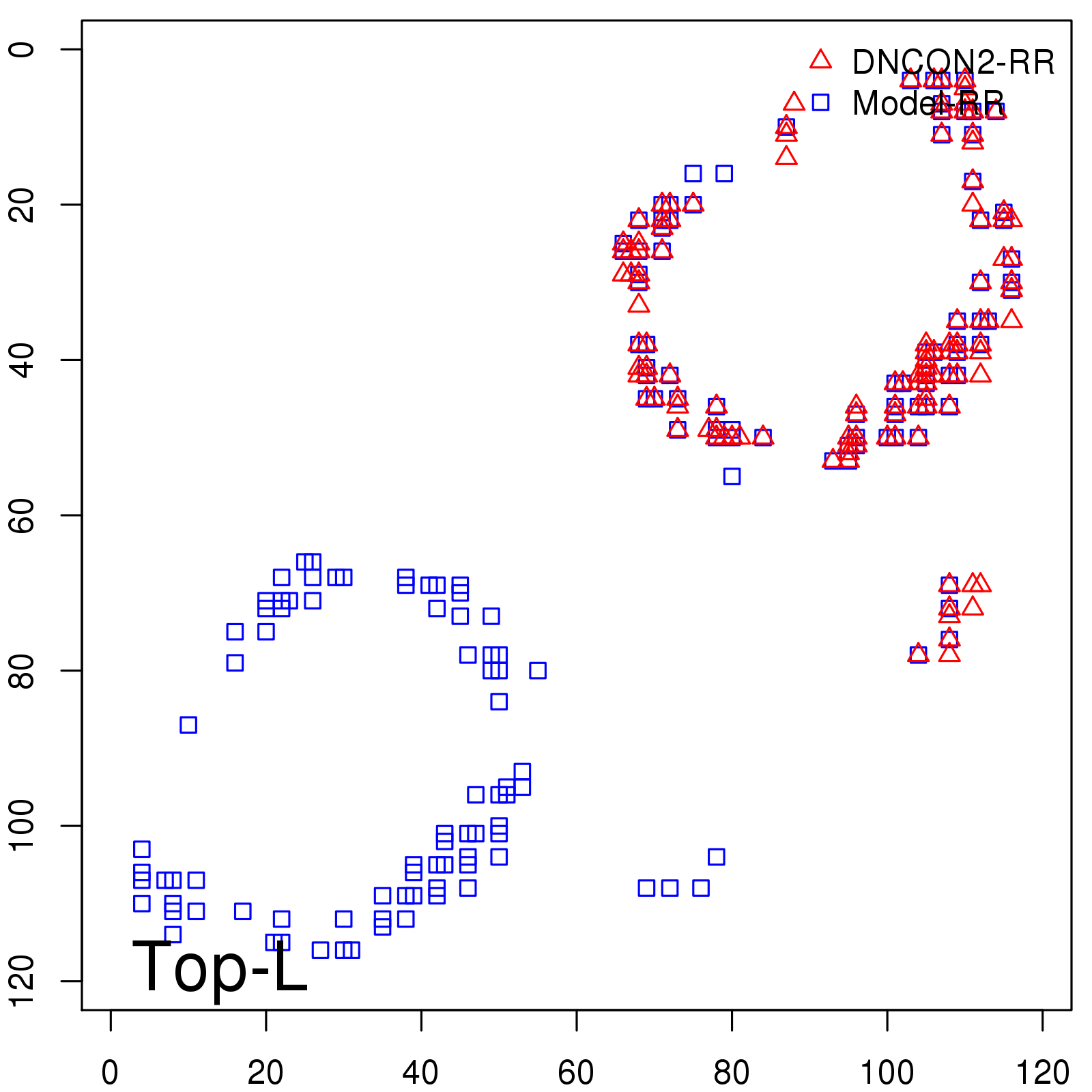
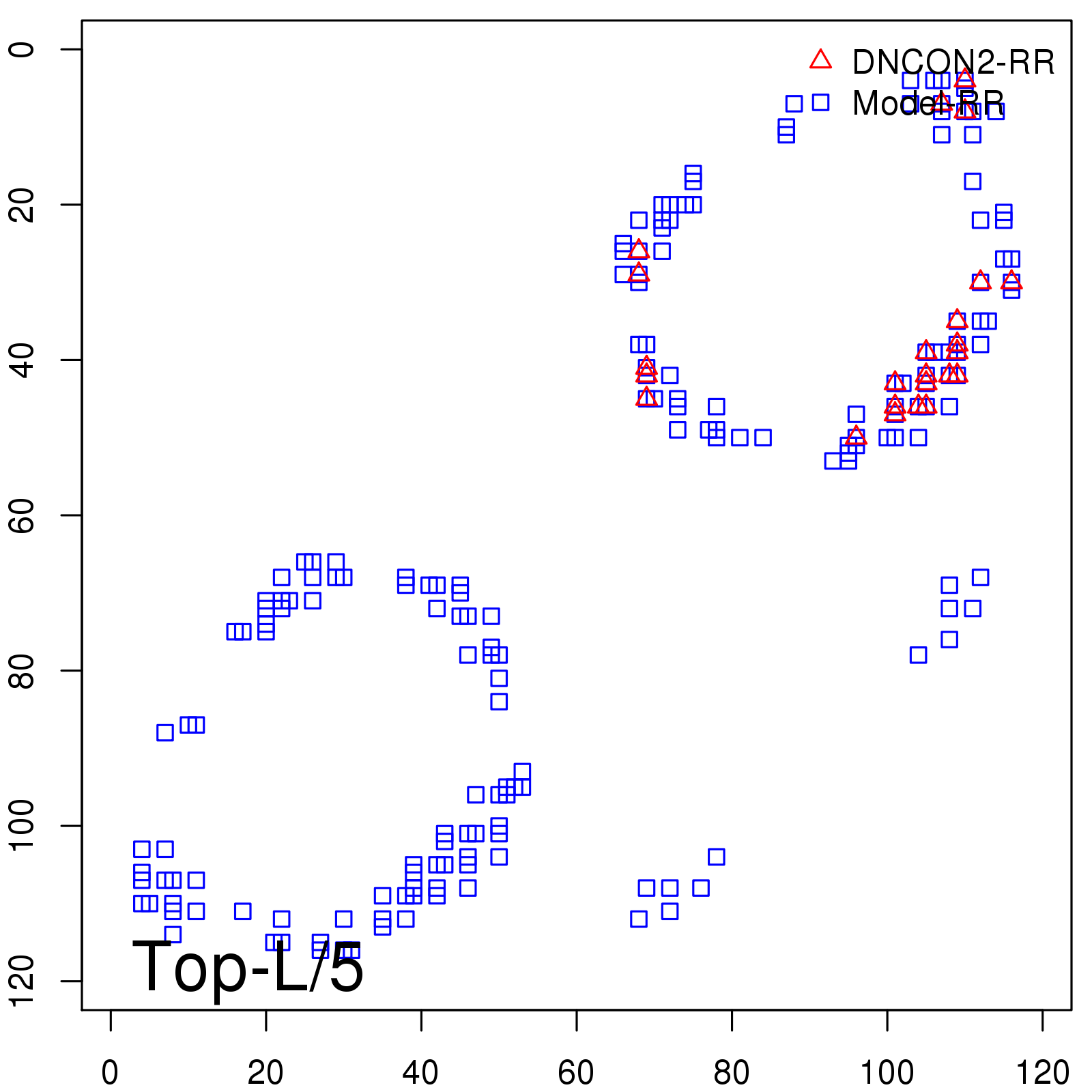
Probability to Precision
|
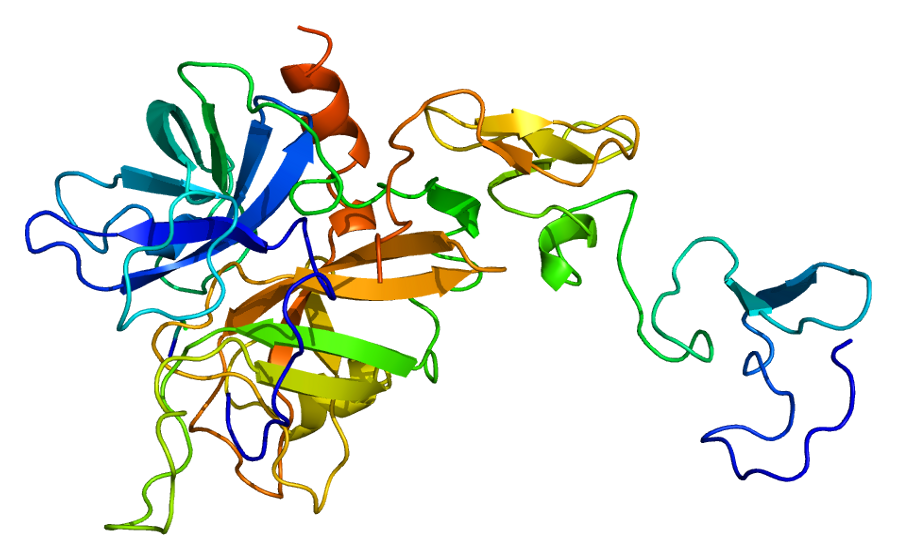
Predicted Top 1 Tertiary structure
|
|
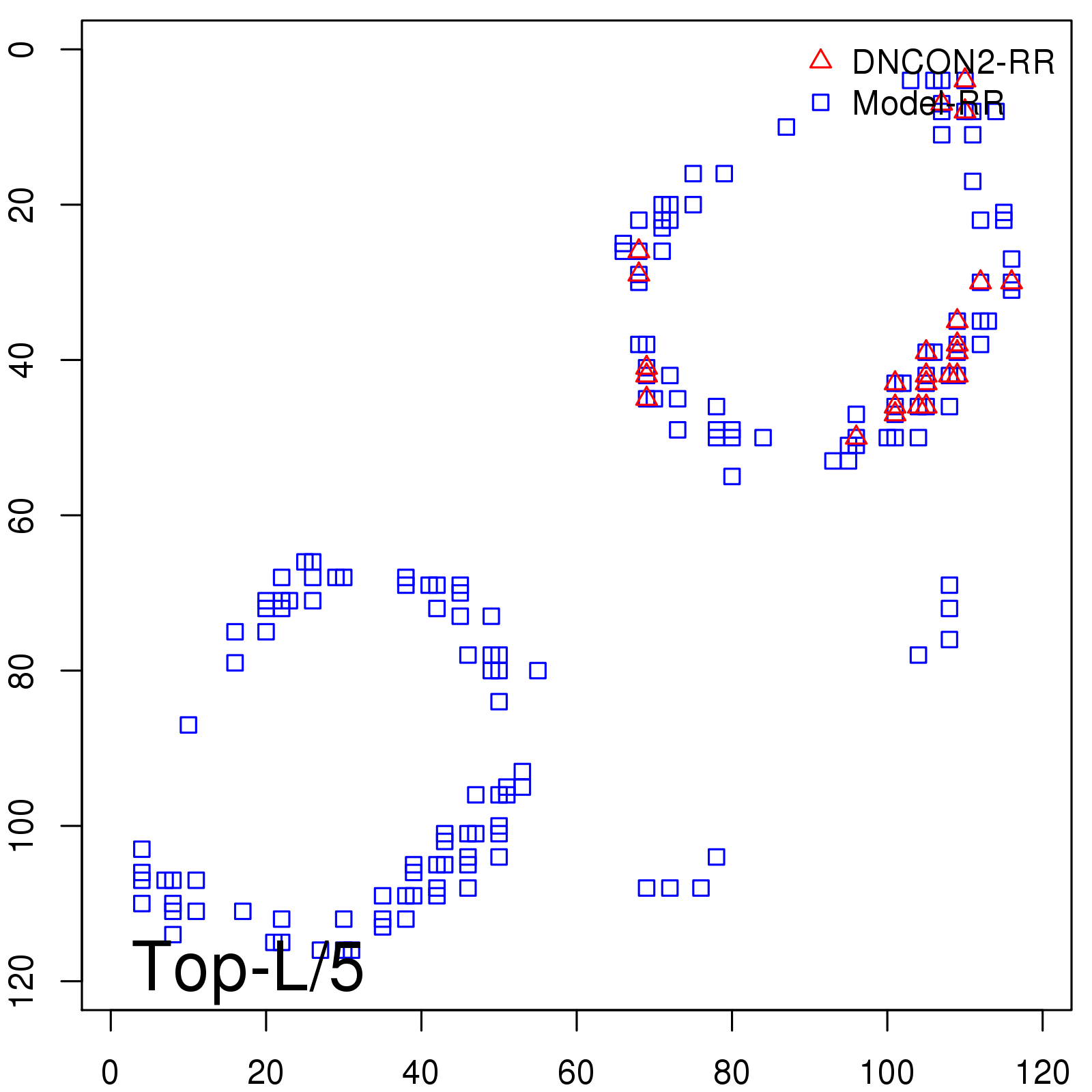
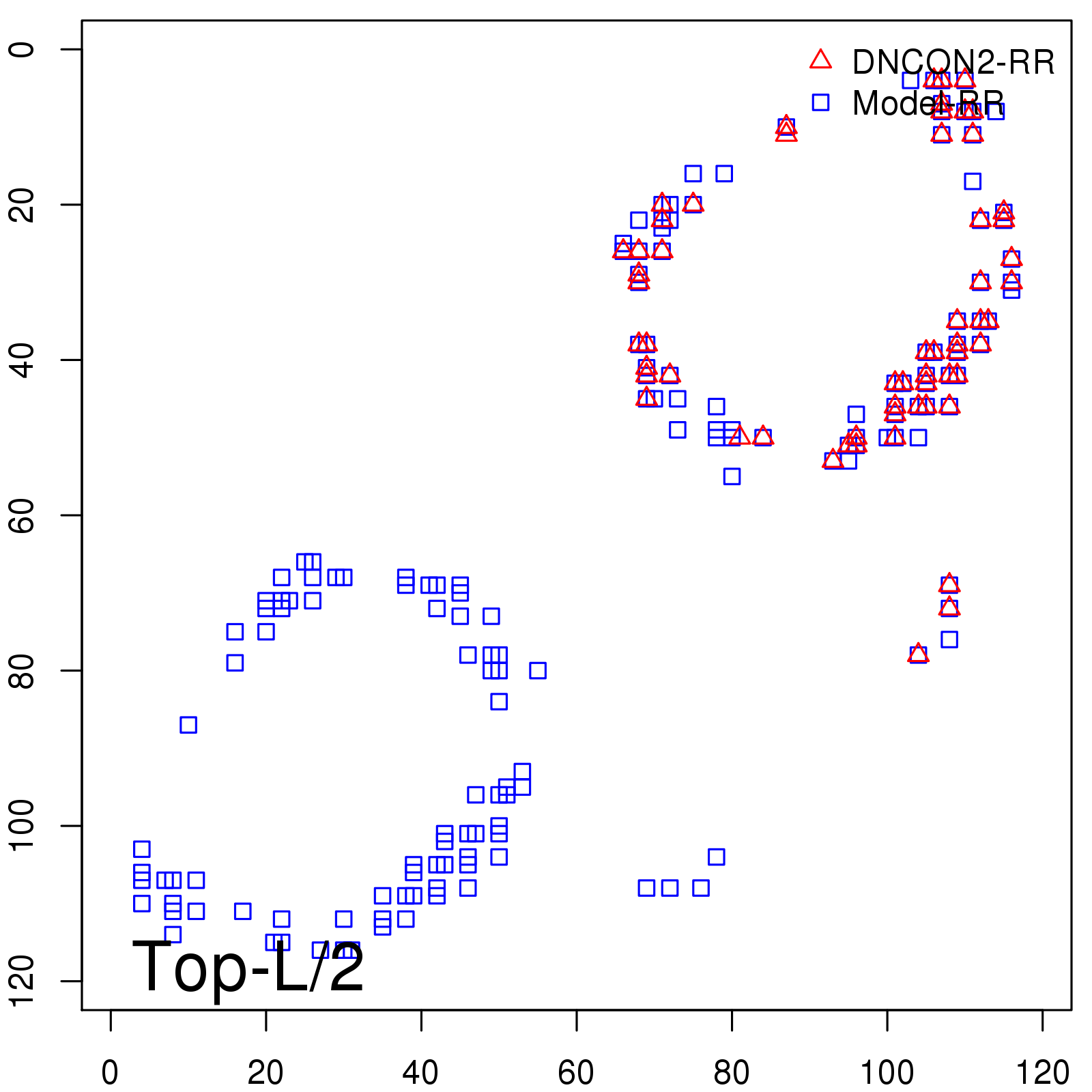
Predicted Contact Accuracy
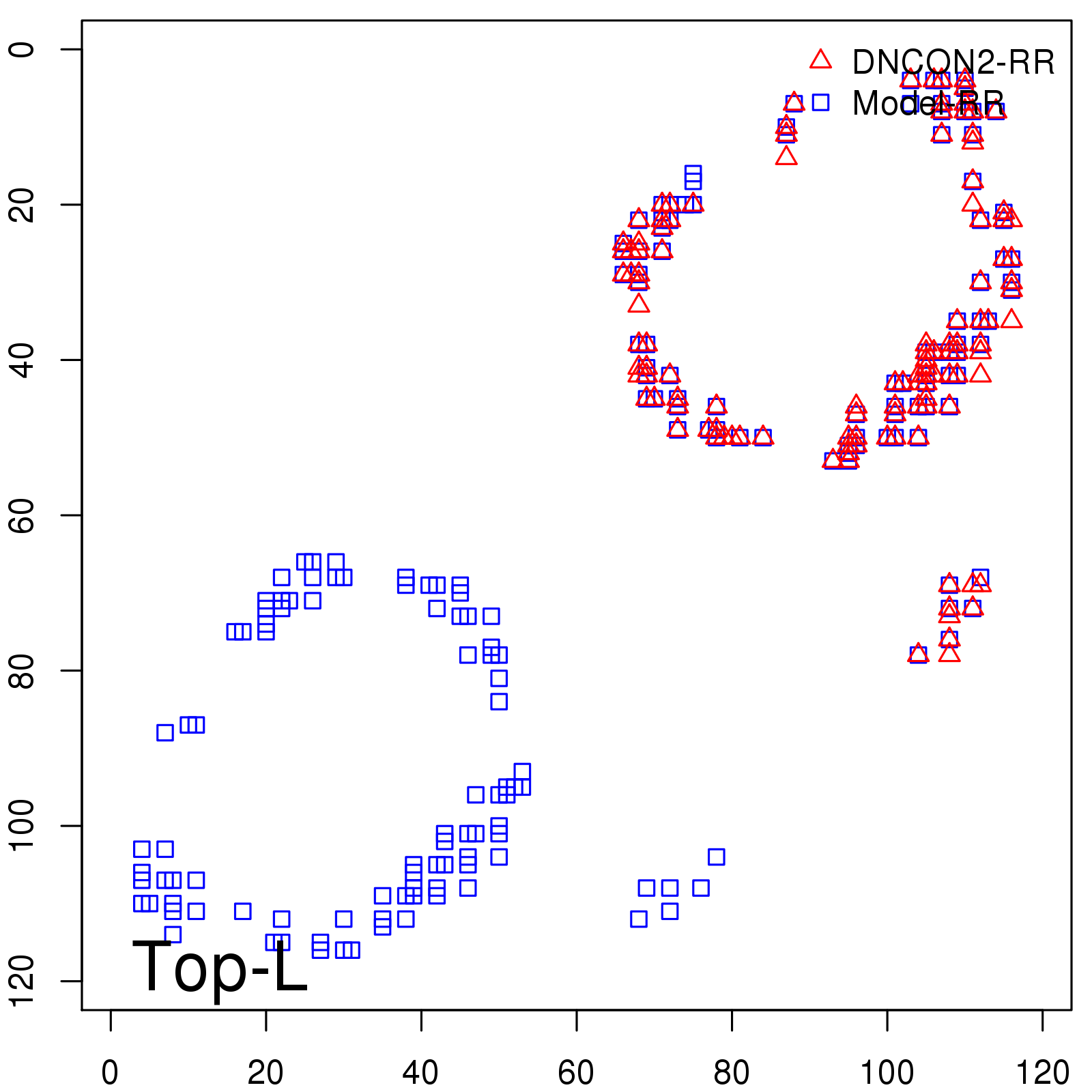
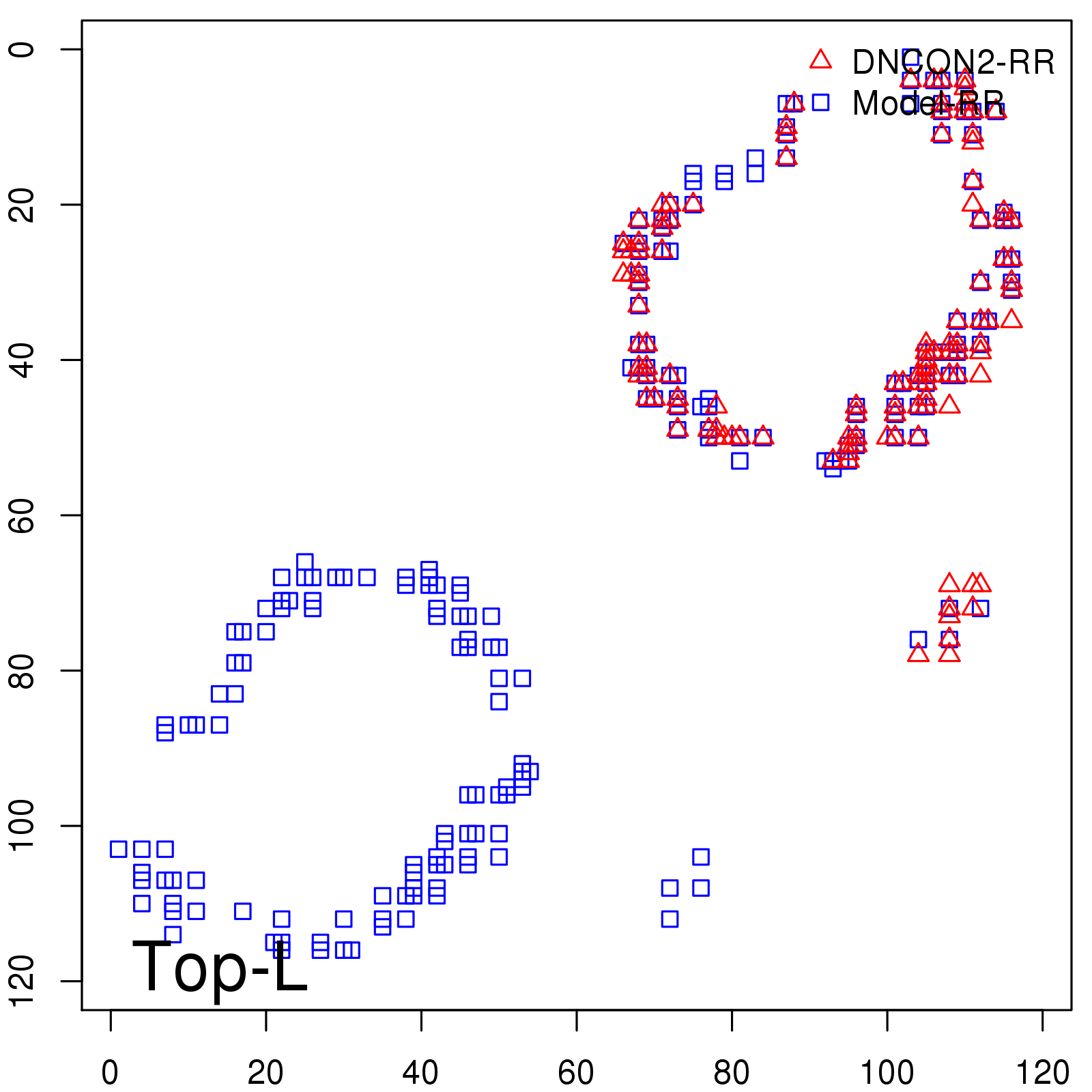
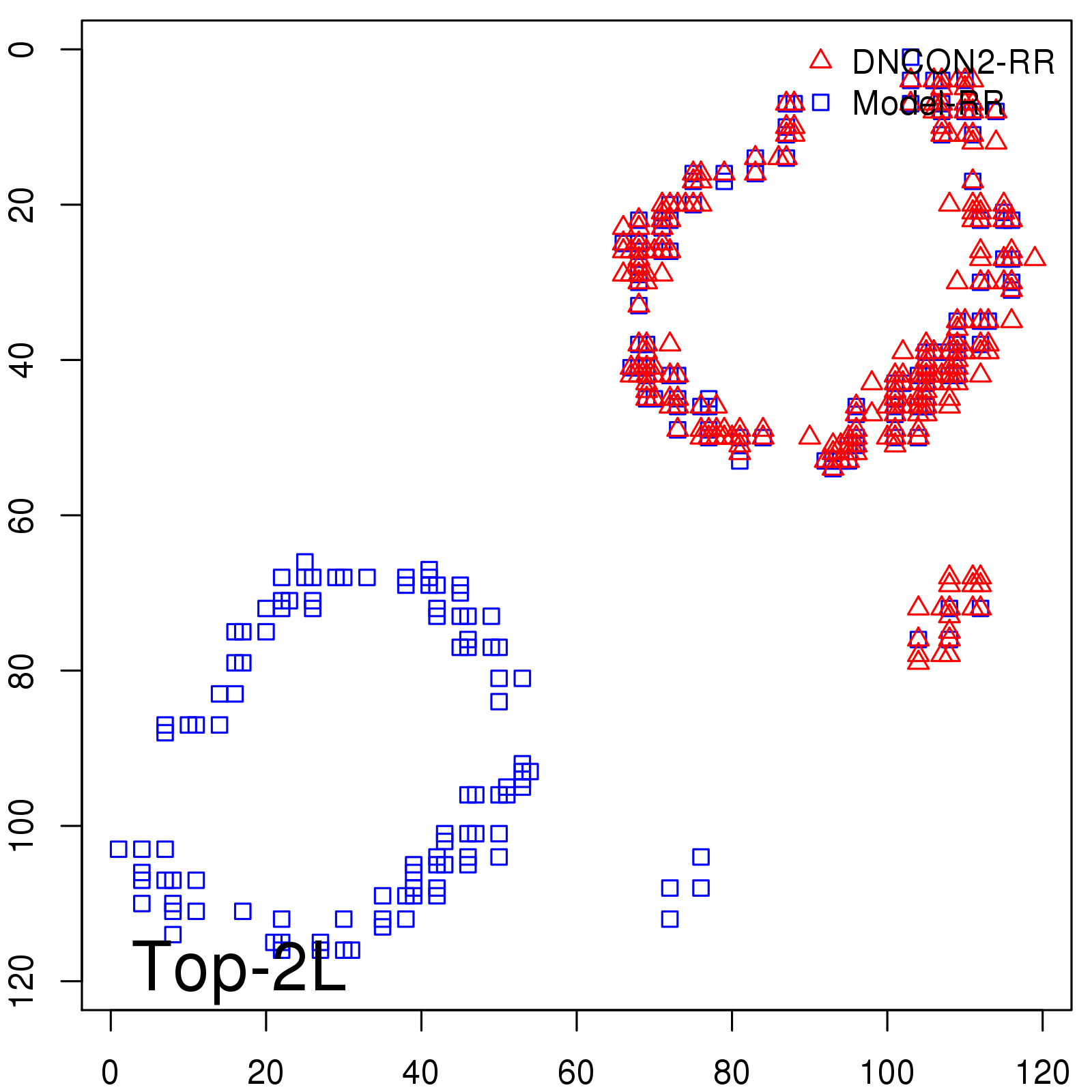
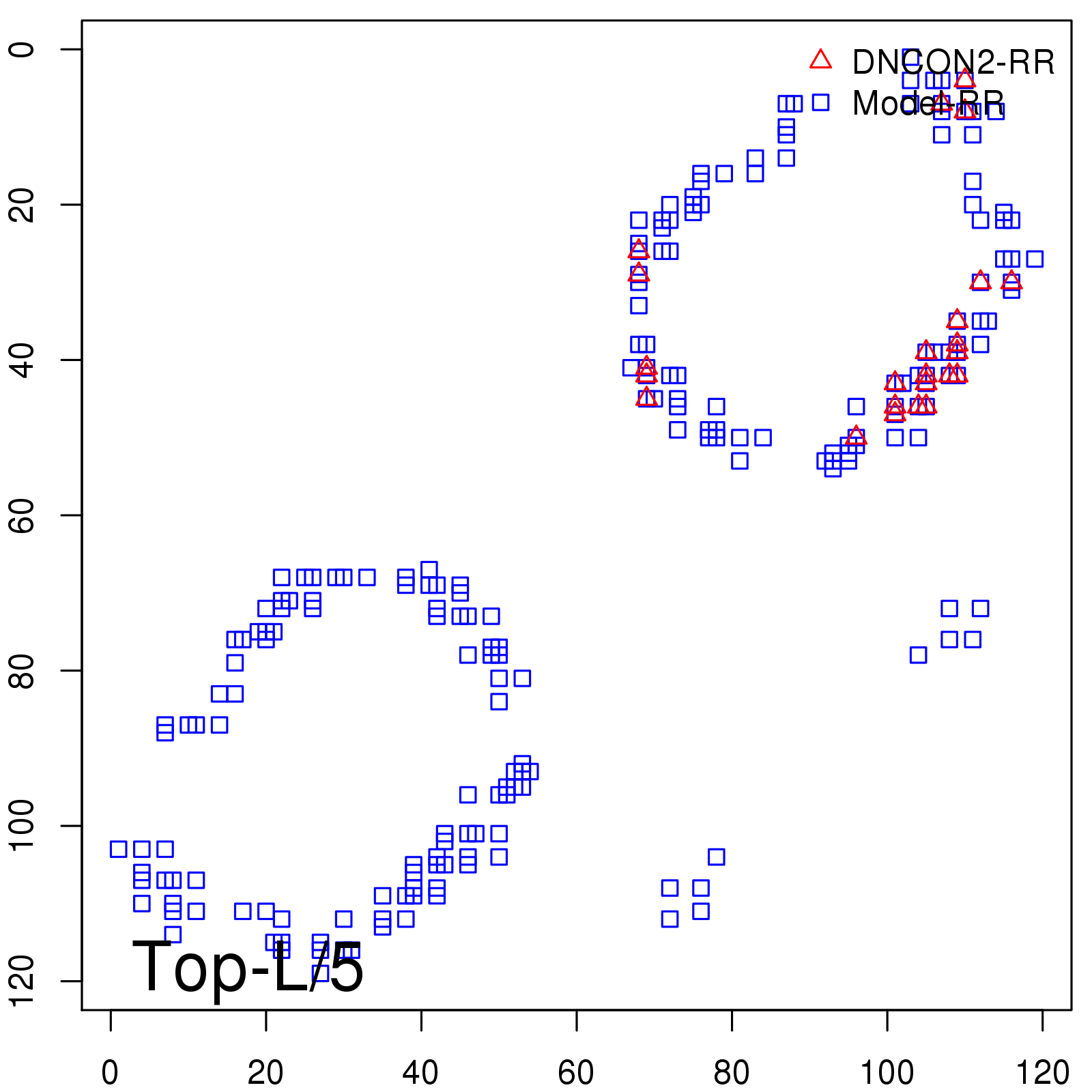
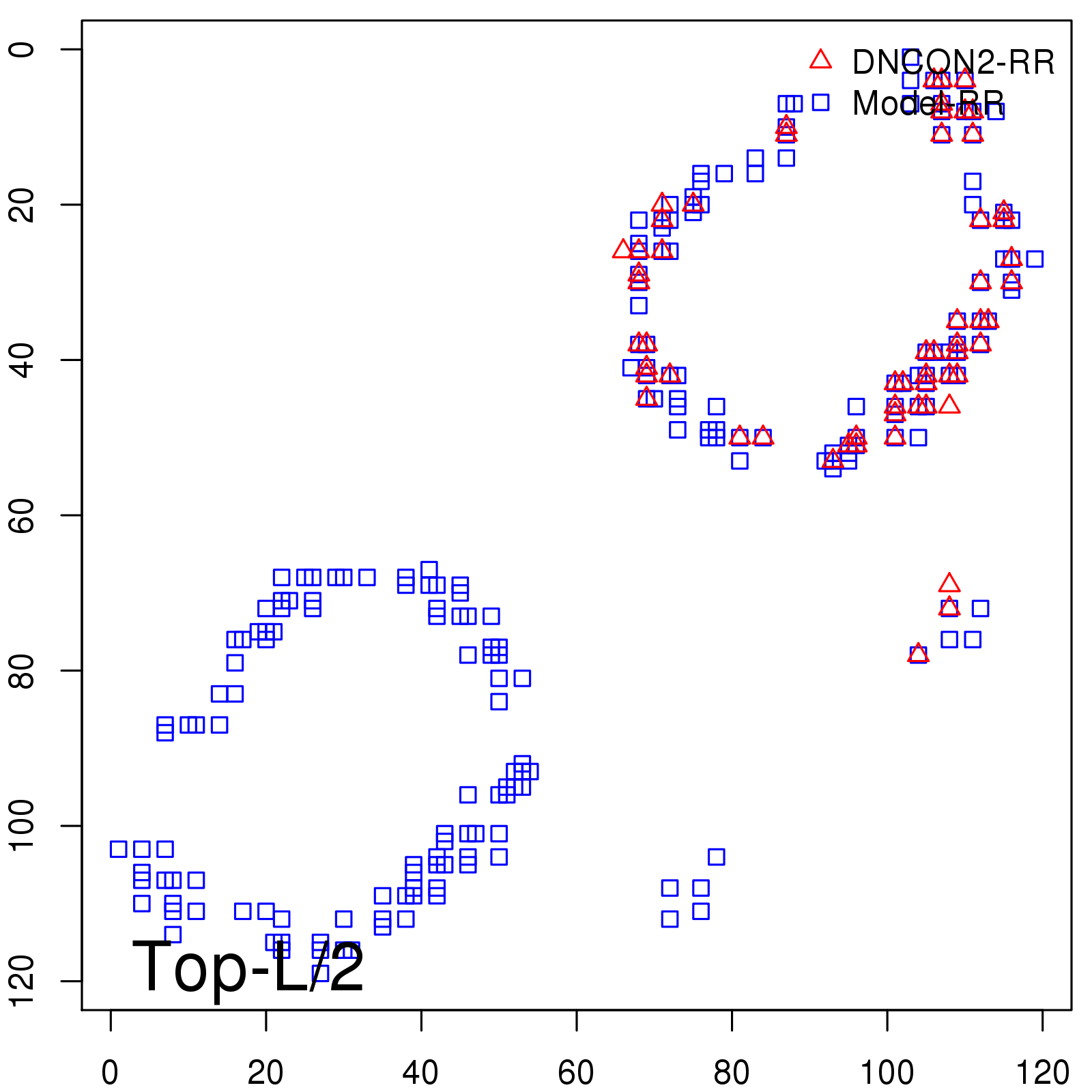
|
| Long-Range | Precision |
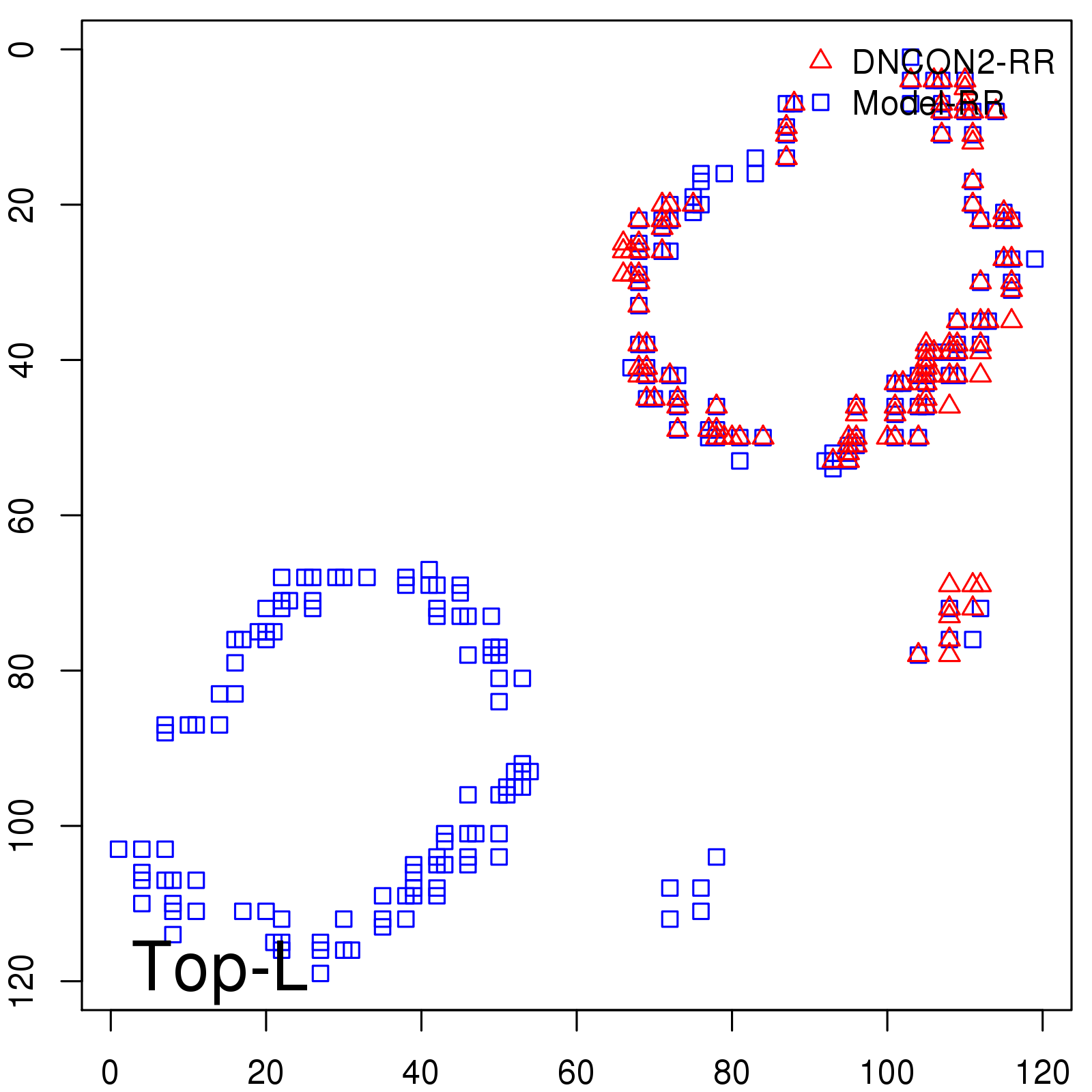
| TopL/5 | 100.00 |
| TopL/2 | 96.67 |
| TopL | 66.39 |
| Top2L | 34.03 |
( Alignment file  )
)
| Alignment | Number |
| N | 1211 |
| Neff | 207 |
Model comparsion
| Model | TM-score |
| Model 2 | 0.9728 |
| Model 3 | 0.8650 |
| Model 4 | 0.8732 |
| Model 5 | 0.8639 |
| Average | 0.89373 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 3tt3 | 0.59664 |
| 6dzz | 0.58283 |
| 2wsx | 0.57312 |
| 3gjc | 0.57082 |
| 6hxm | 0.56560 |
Predicted Top 2 Tertiary structure
|
|
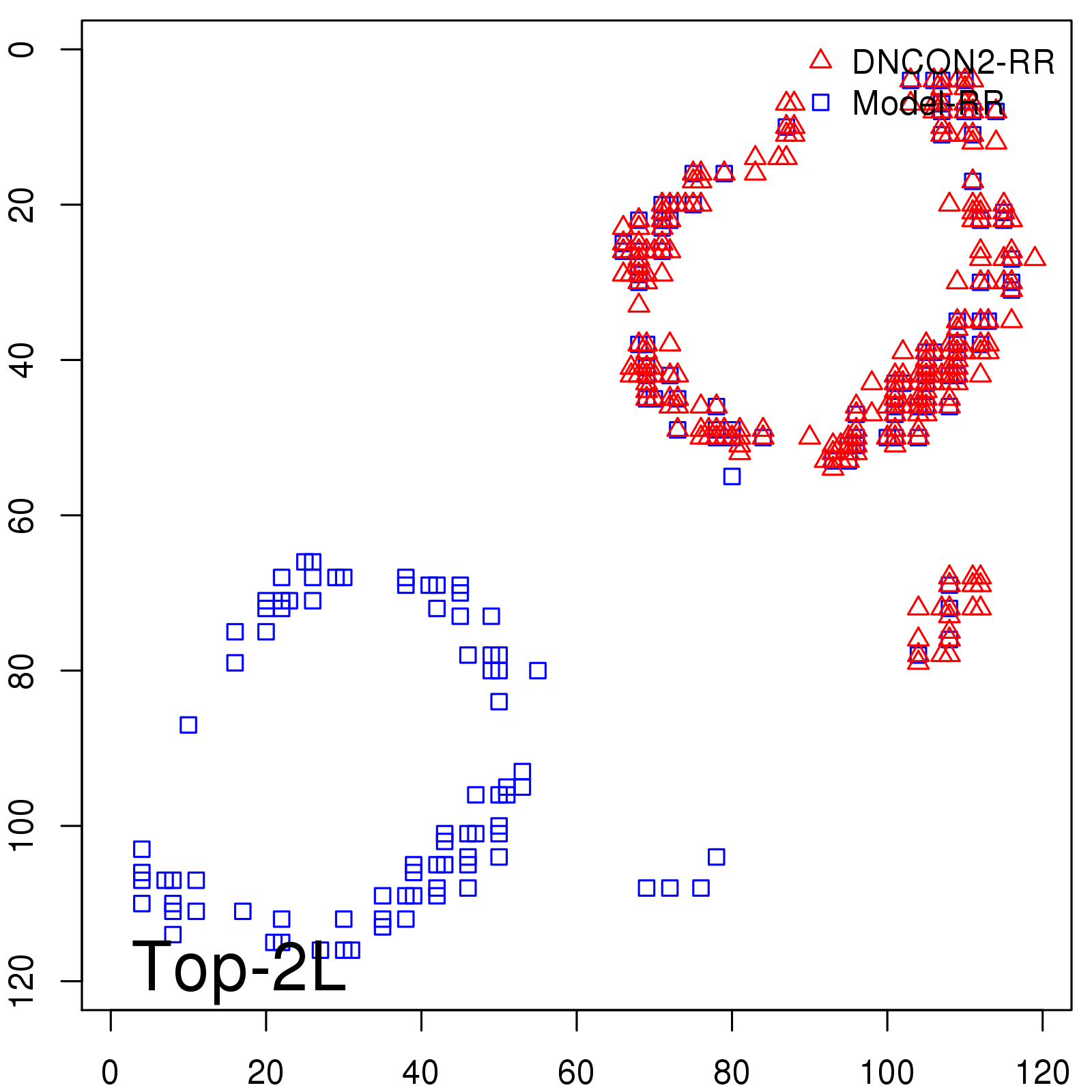
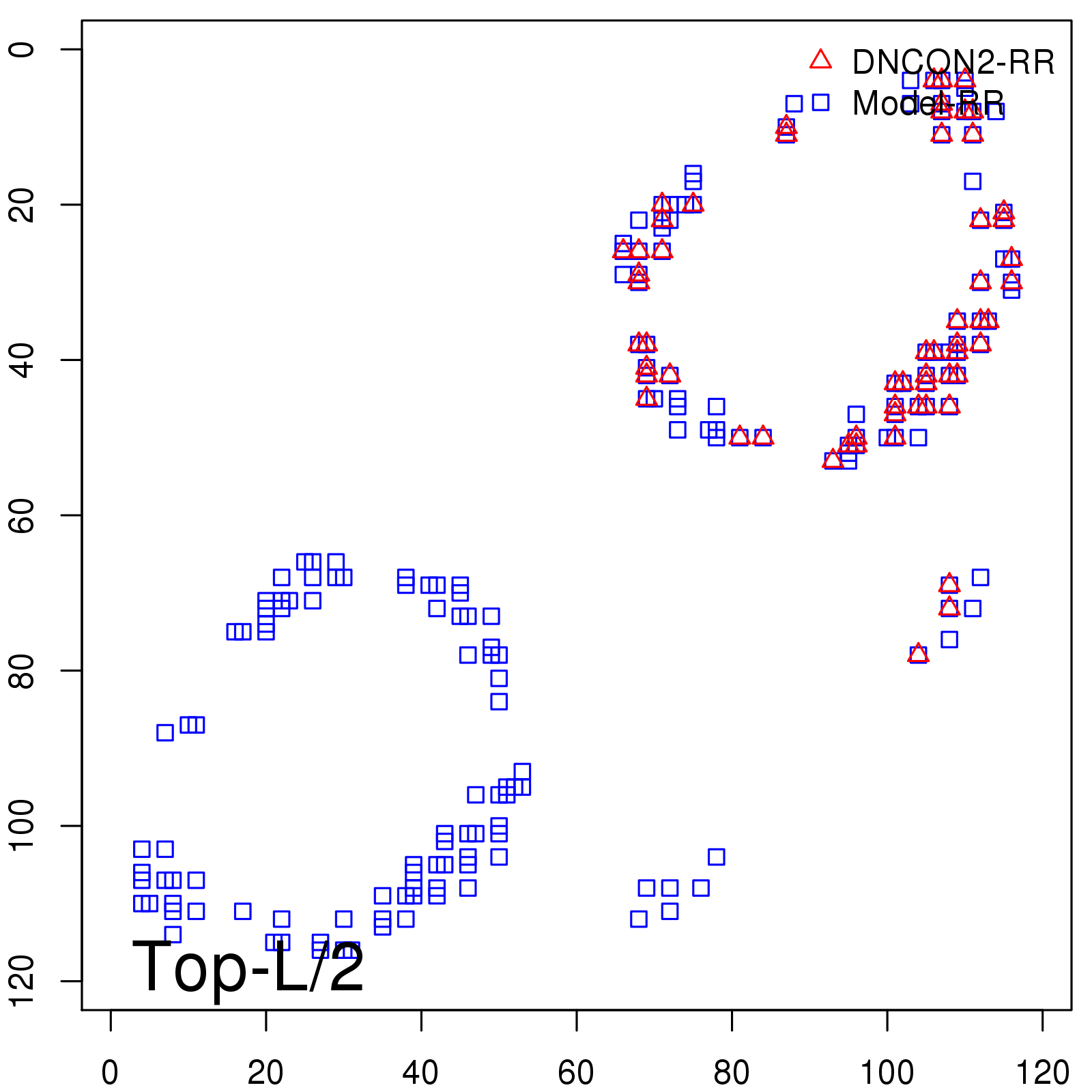
Predicted Contact Accuracy
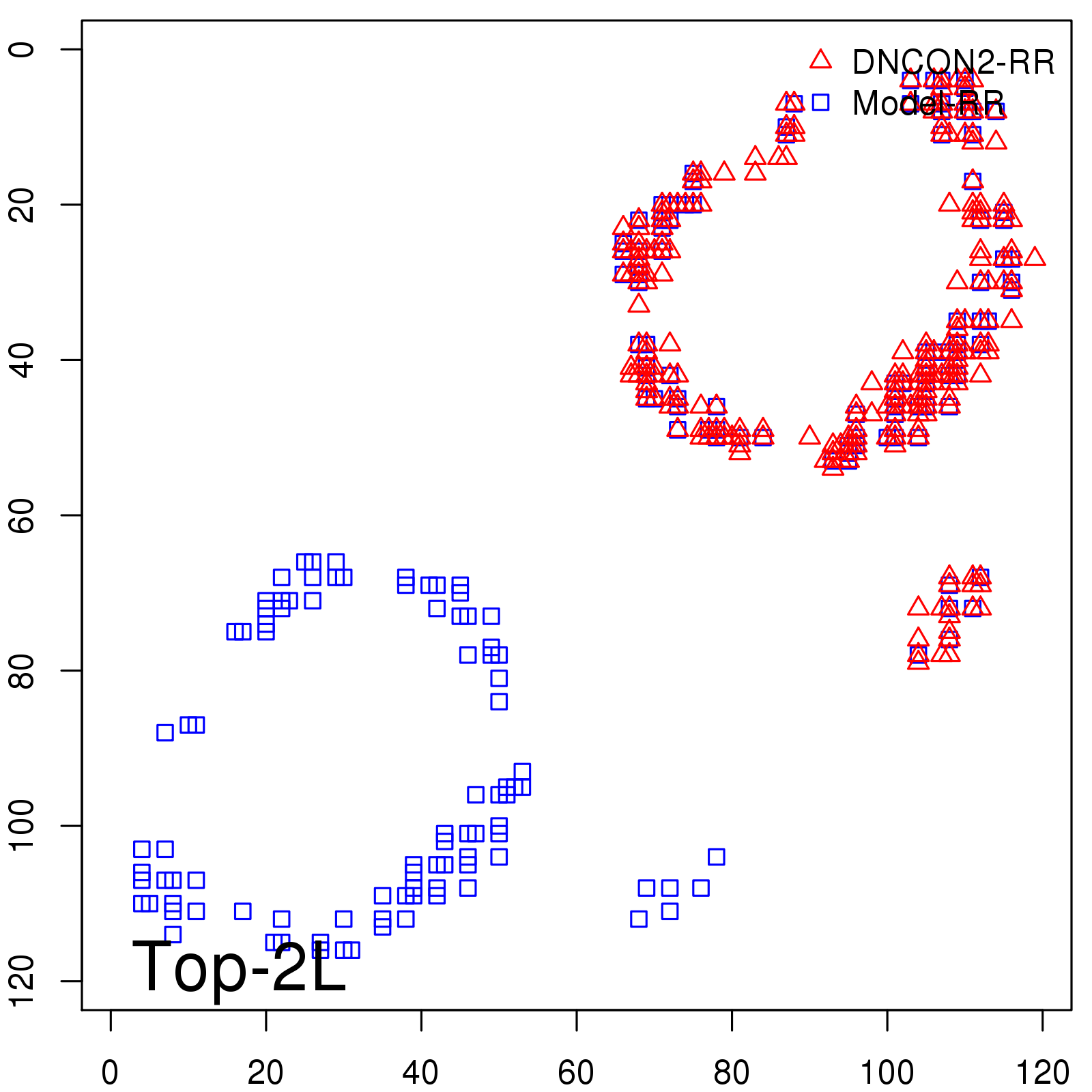
|
| Long-Range | Precision |
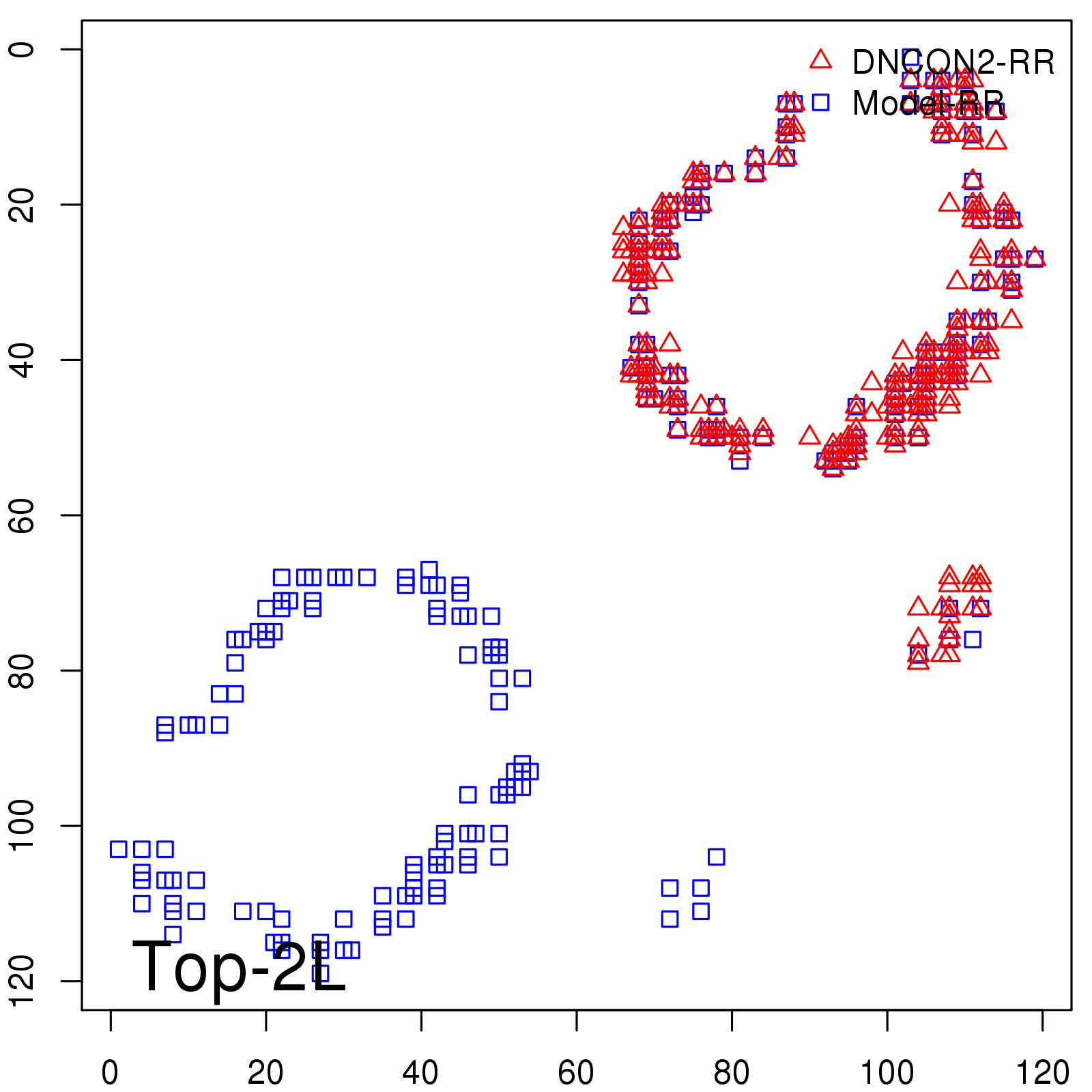
| TopL/5 | 100.00 |
| TopL/2 | 100.00 |
| TopL | 74.79 |
| Top2L | 39.50 |
( Alignment file  )
)
| Alignment | Number |
| N | 1211 |
| Neff | 207 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9728 |
| Model 3 | 0.8806 |
| Model 4 | 0.8865 |
| Model 5 | 0.8789 |
| Average | 0.90470 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 3tt3 | 0.59599 |
| 3sf5 | 0.58582 |
| 3gjc | 0.56997 |
| 6hxm | 0.56281 |
| 6hxl | 0.55602 |
Predicted Top 3 Tertiary structure
|
|
Predicted Contact Accuracy
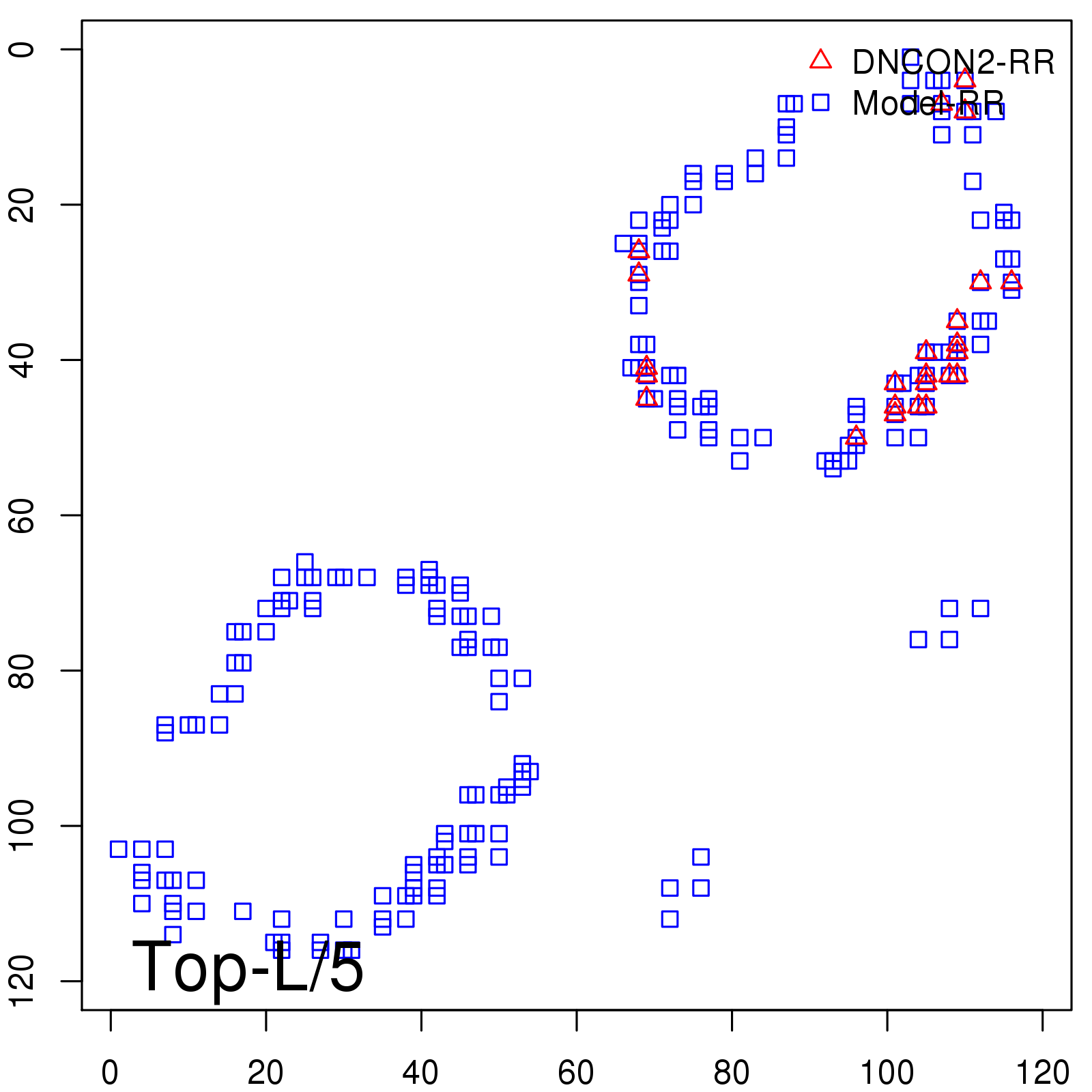
|
| Long-Range | Precision |
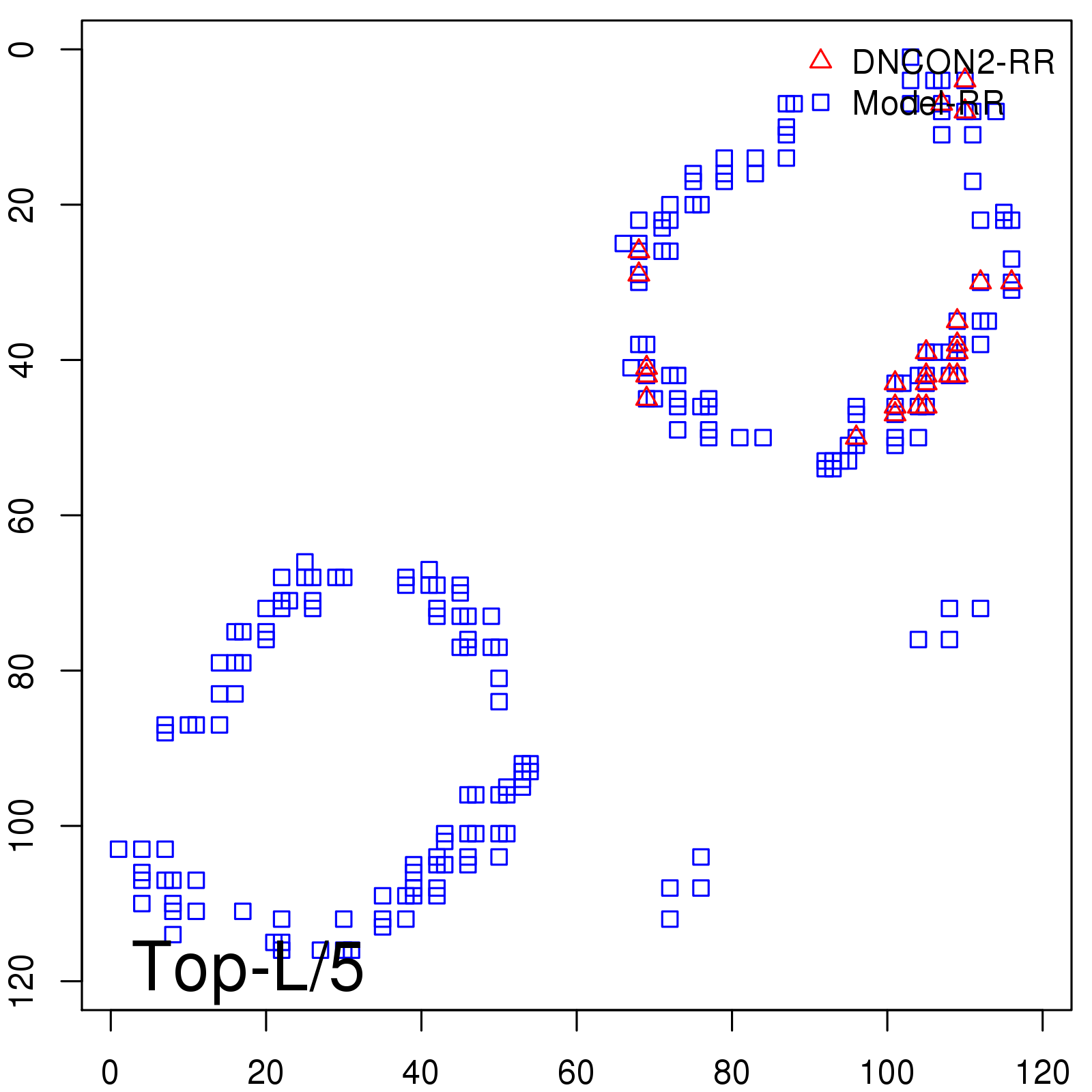
| TopL/5 | 100.00 |
| TopL/2 | 91.67 |
| TopL | 69.75 |
| Top2L | 42.02 |
( Alignment file  )
)
| Alignment | Number |
| N | 1211 |
| Neff | 207 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.8650 |
| Model 2 | 0.8806 |
| Model 4 | 0.9714 |
| Model 5 | 0.9839 |
| Average | 0.92522 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 6jc4 | 0.57470 |
| 3sf5 | 0.56651 |
| 3tt3 | 0.56600 |
| 5xwp | 0.56465 |
| 6hxm | 0.55723 |
Predicted Top 4 Tertiary structure
|
|
Predicted Contact Accuracy
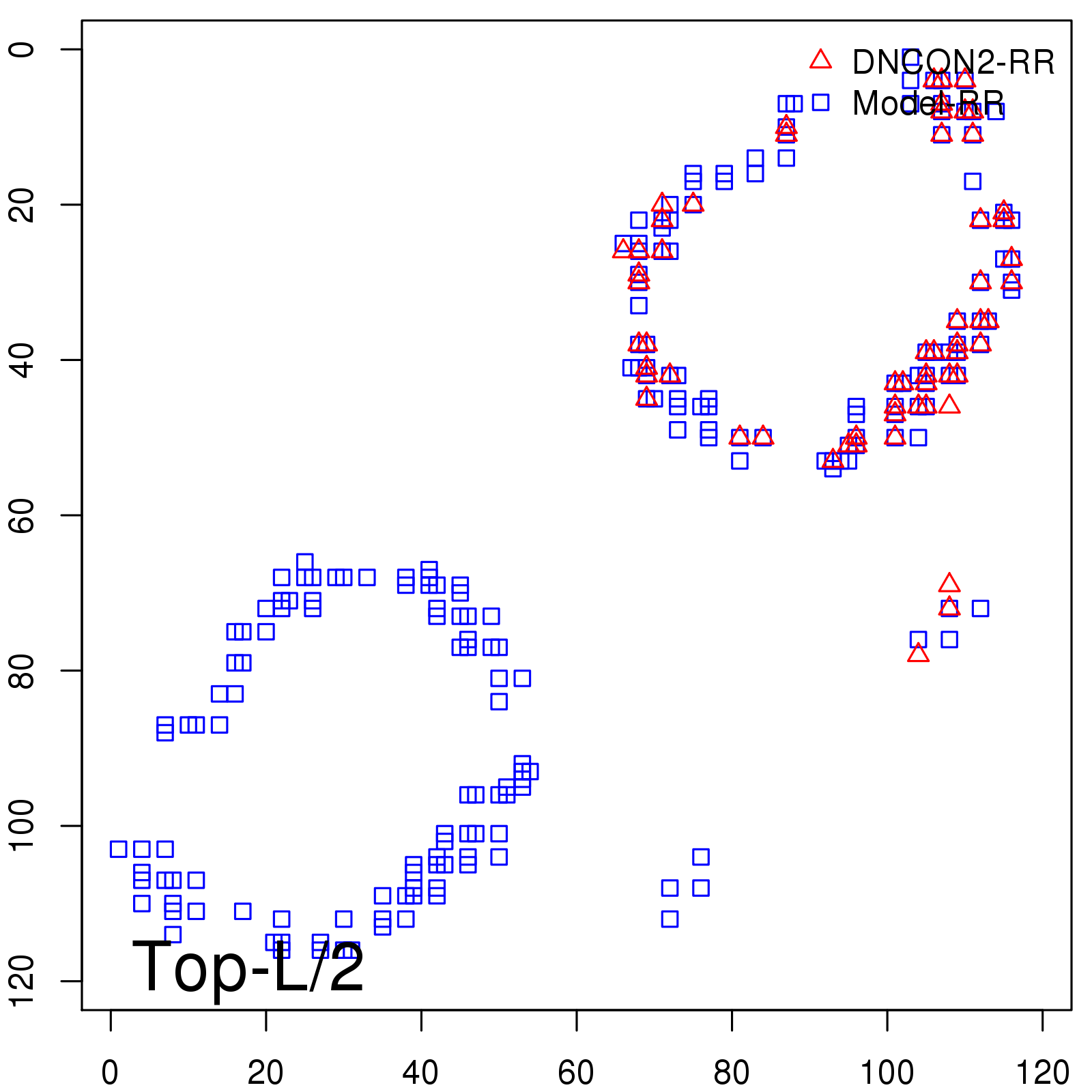
|
| Long-Range | Precision |
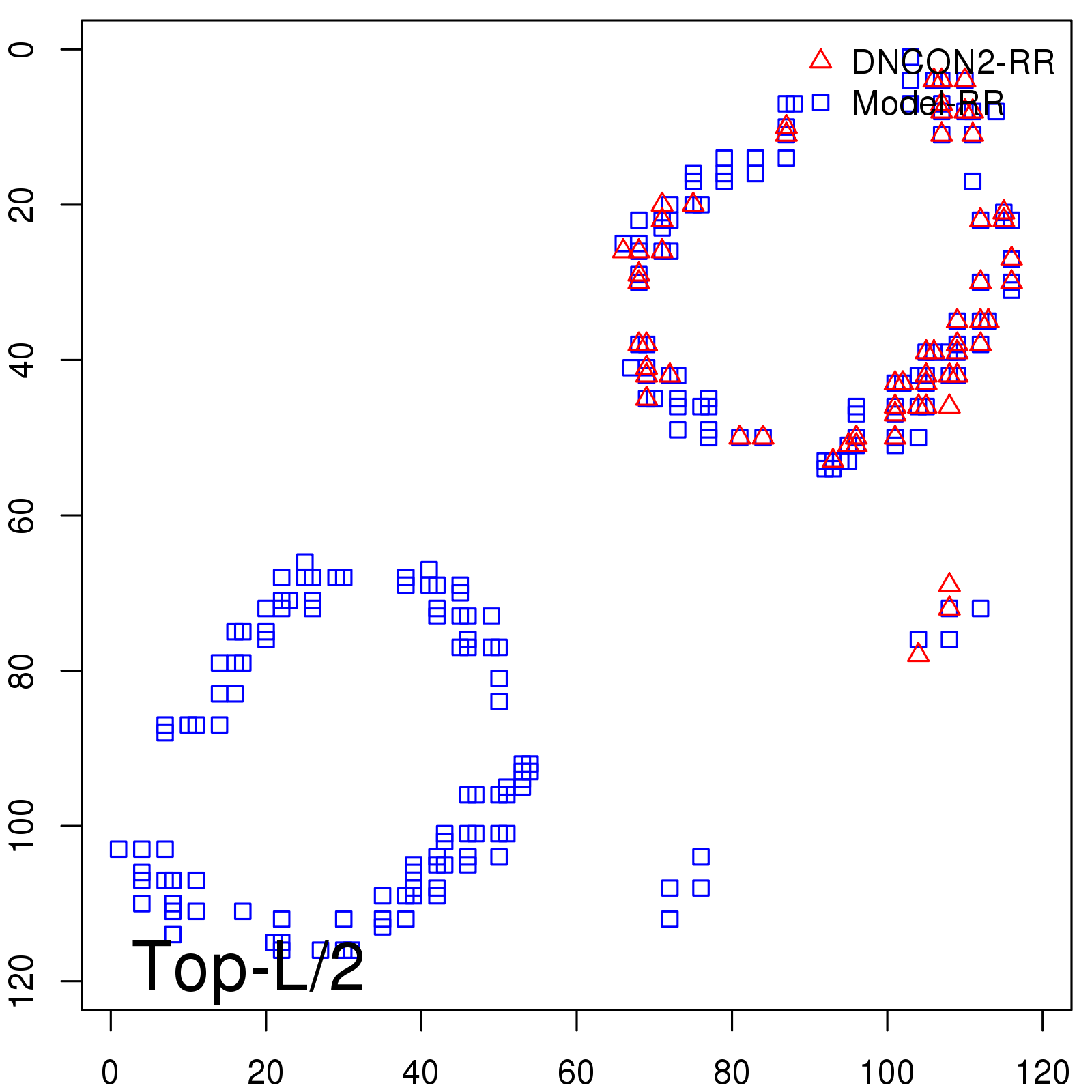
| TopL/5 | 100.00 |
| TopL/2 | 93.33 |
| TopL | 72.27 |
| Top2L | 43.28 |
( Alignment file  )
)
| Alignment | Number |
| N | 1211 |
| Neff | 207 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.8732 |
| Model 2 | 0.8865 |
| Model 3 | 0.9714 |
| Model 5 | 0.9750 |
| Average | 0.92653 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 3tt3 | 0.57034 |
| 6jc4 | 0.57011 |
| 3sf5 | 0.56808 |
| 5xwp | 0.56450 |
| 3hfx | 0.56375 |
Predicted Top 5 Tertiary structure
|
Jmol._Canvas2D (Jmol) "jsmolApplet_M23d5"[x] spinFPS is set too fast (30) -- can't keep up! spinFPS is set too fast (30) -- can't keep up! spinFPS is set too fast (30) -- can't keep up! |
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 91.67 |
| TopL | 67.23 |
| Top2L | 41.60 |
( Alignment file  )
)
| Alignment | Number |
| N | 1211 |
| Neff | 207 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.8639 |
| Model 2 | 0.8789 |
| Model 3 | 0.9839 |
| Model 4 | 0.9750 |
| Average | 0.92543 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 3tt3 | 0.56943 |
| 6jc4 | 0.56876 |
| 4hi0 | 0.56434 |
| 3sf5 | 0.56147 |
| 5xwp | 0.56098 |