Results of Structure Prediction for Target Name: WP_010940344 ( Click  )
)
Domain Boundary prediction ( View  )
)

Protein sequence
| 1-60: | M | K | Y | I | I | F | E | D | F | A | G | H | P | E | P | F | L | F | P | R | R | V | D | H | G | D | M | R | E | Q | L | P | Y | A | K | V | L | S | A | G | Y | V | H | F | S | G | G | V | F | T | C | H | G | G | S | A | E | L | D | T |
| 61-81: | V | A | R | P | E | D | A | D | I | I | A | T | R | L | A | P | R | D | T | G | V |
Secondary structure prediction (H: Helix E: Strand C: Coil)
| 1-60: | C | C | E | E | E | E | E | E | C | C | C | C | C | C | C | E | E | C | C | C | C | C | C | H | H | H | H | H | H | H | C | C | C | H | H | E | E | E | C | C | E | E | E | E | C | C | C | E | E | E | E | C | C | C | C | H | H | H | H | H |
| 61-81: | H | C | C | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C |
| H(Helix): 26(32.1%) | E(Strand): 19(23.46%) | C(Coil): 36(44.44%) |
Solvent accessibility prediction (e: Exposed b: Buried)
| 1-60: | E | E | B | B | B | B | E | B | B | E | E | E | E | E | B | B | B | B | B | E | E | B | E | E | E | E | B | E | E | E | B | E | B | B | E | B | B | B | B | B | B | B | B | B | E | E | E | E | B | E | B | E | B | E | B | E | E | B | E | B |
| 61-81: | E | B | E | E | E | B | B | E | B | B | B | E | E | B | E | E | E | E | E | E | E |
| e(Exposed): 43(53.09%) | b(Buried): 38(46.91%) |
Disorder prediction (N: Normal T: Disorder)
| 1-60: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 61-81: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T | T |
| N(Normal): 77(95.06%) | T(Disorder): 4(4.94%) |
Select domain
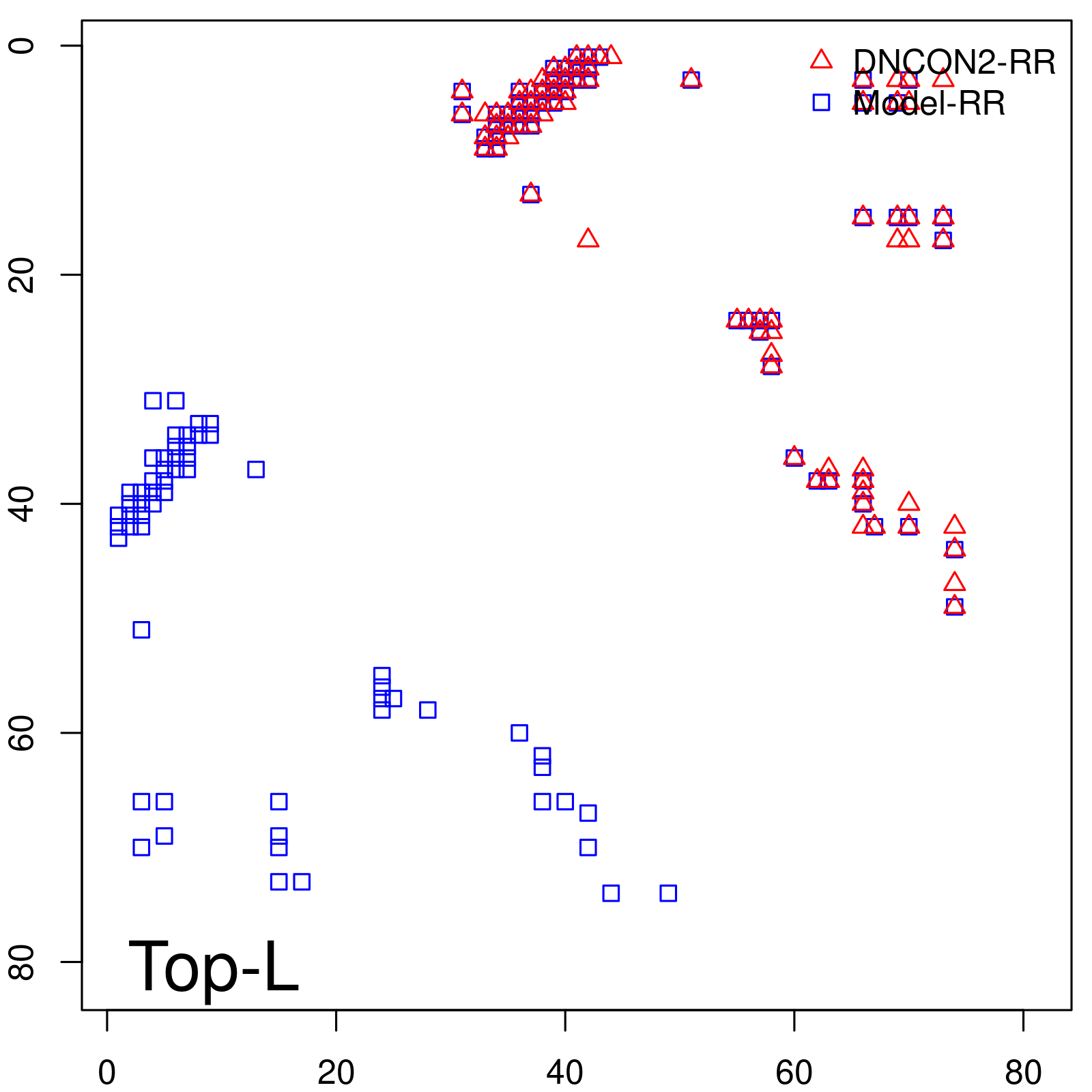
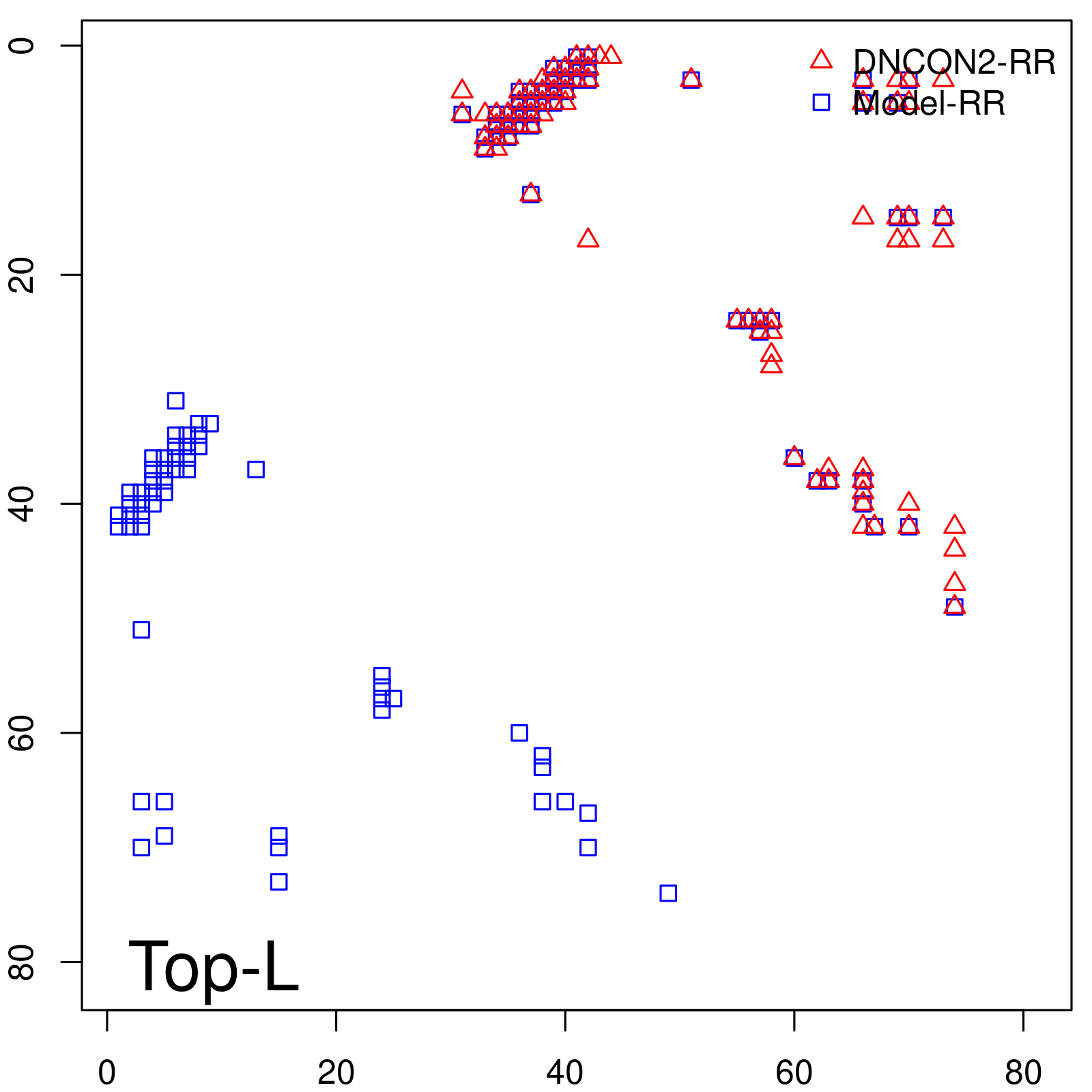
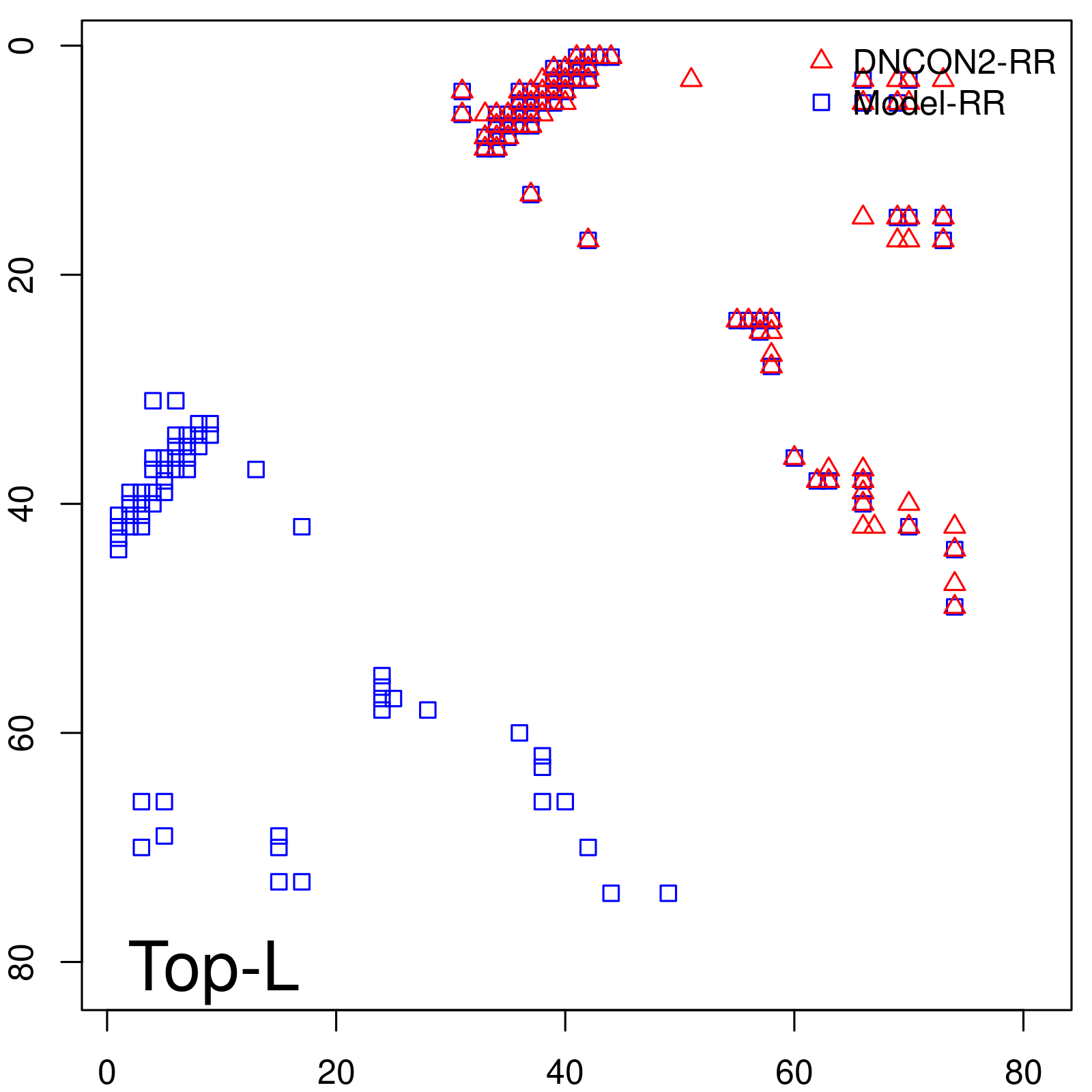
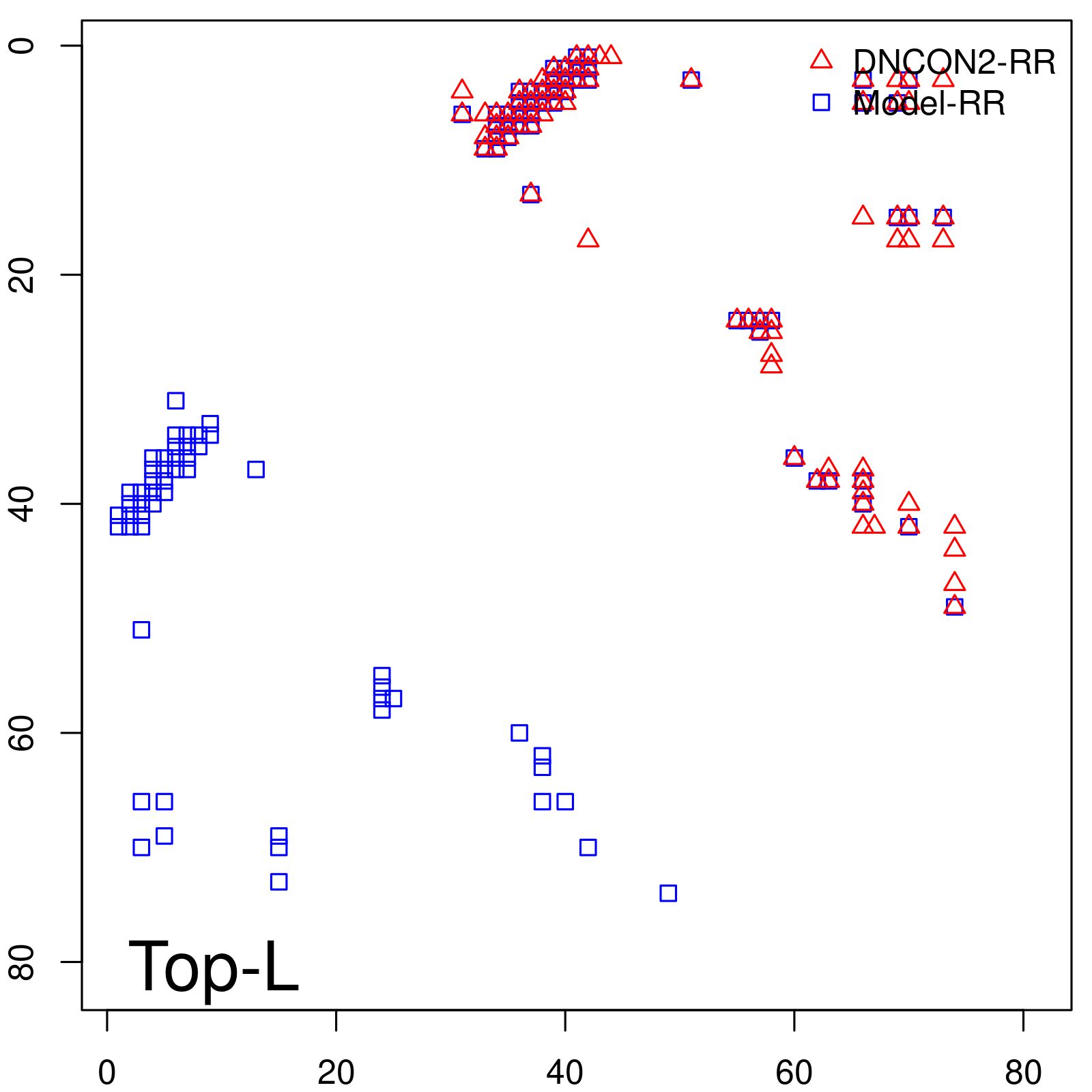
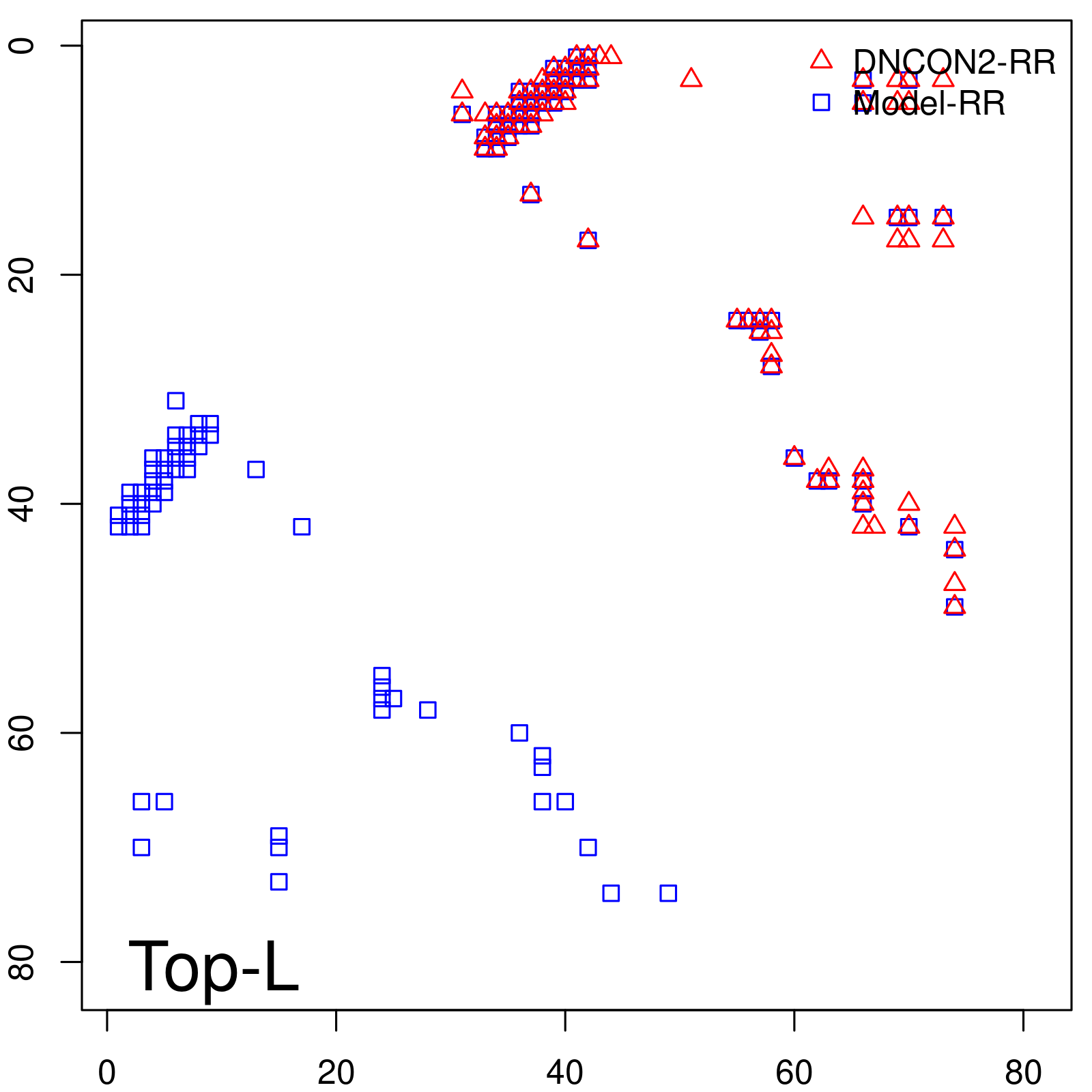
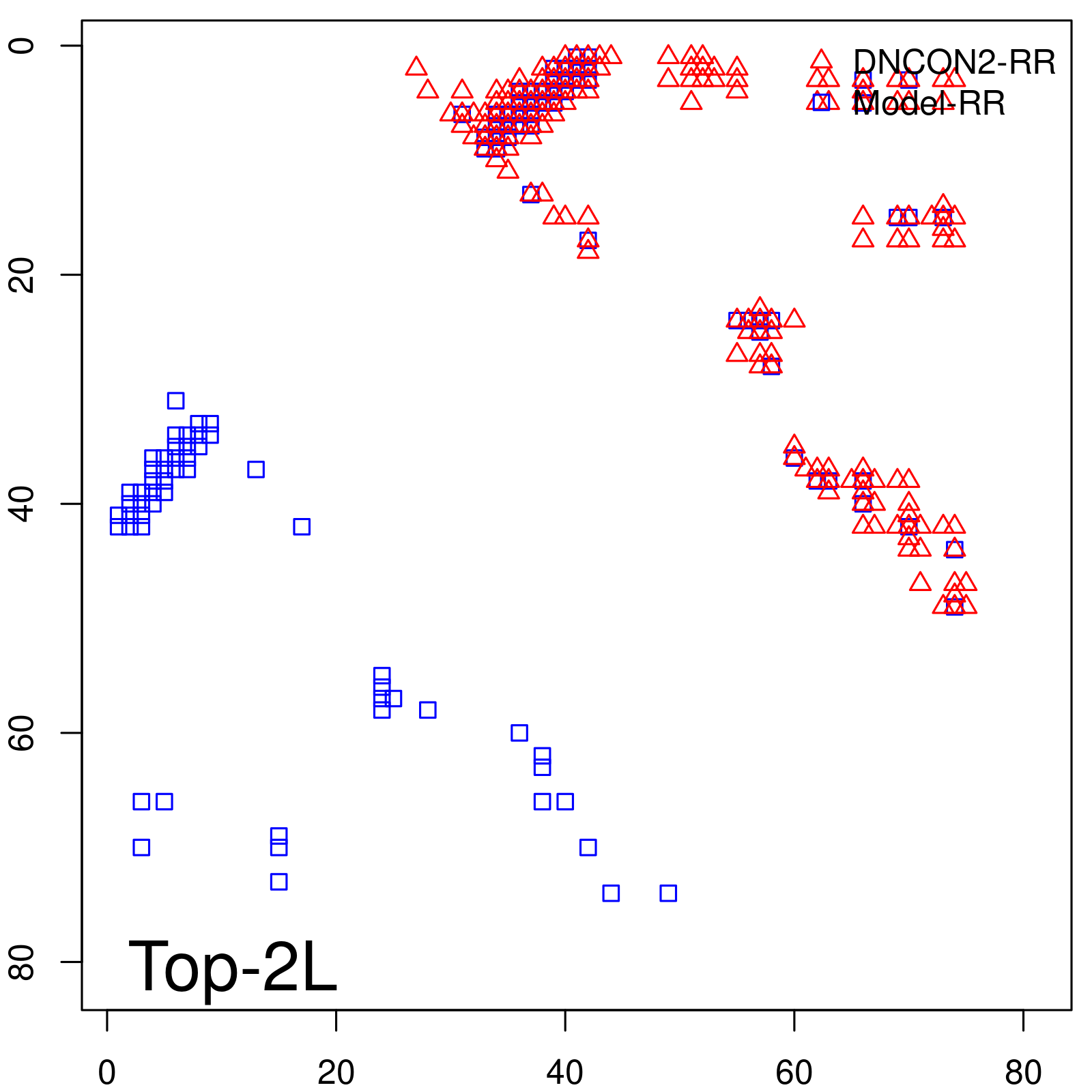
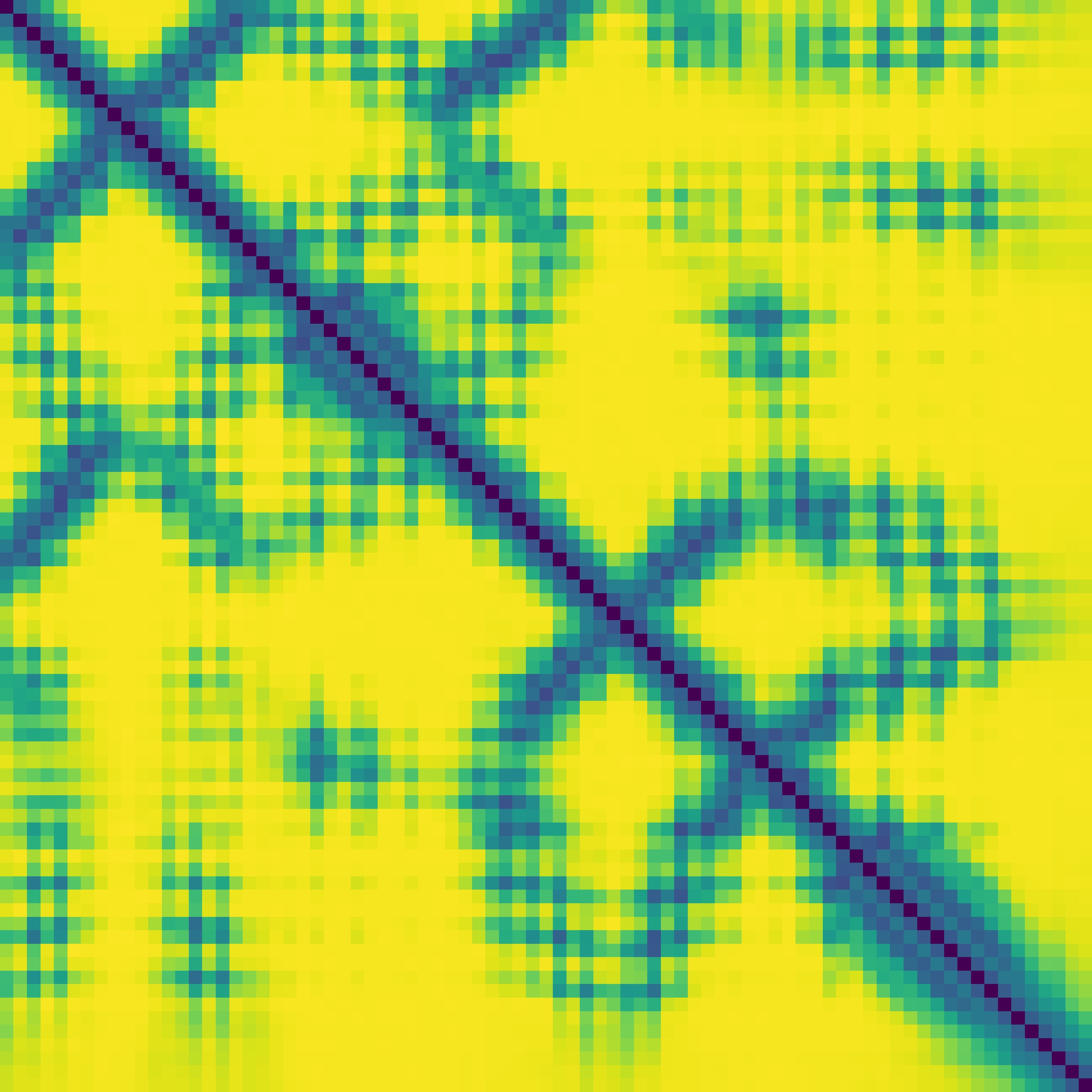
Predicted contact map and distance map
|
|

|
Probability to Precision |
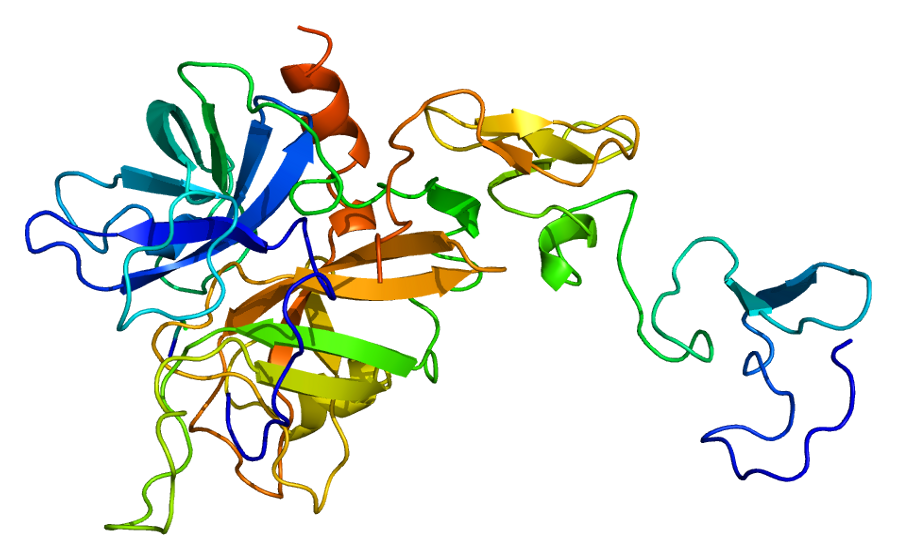
Predicted Top 1 Tertiary structure
|
|
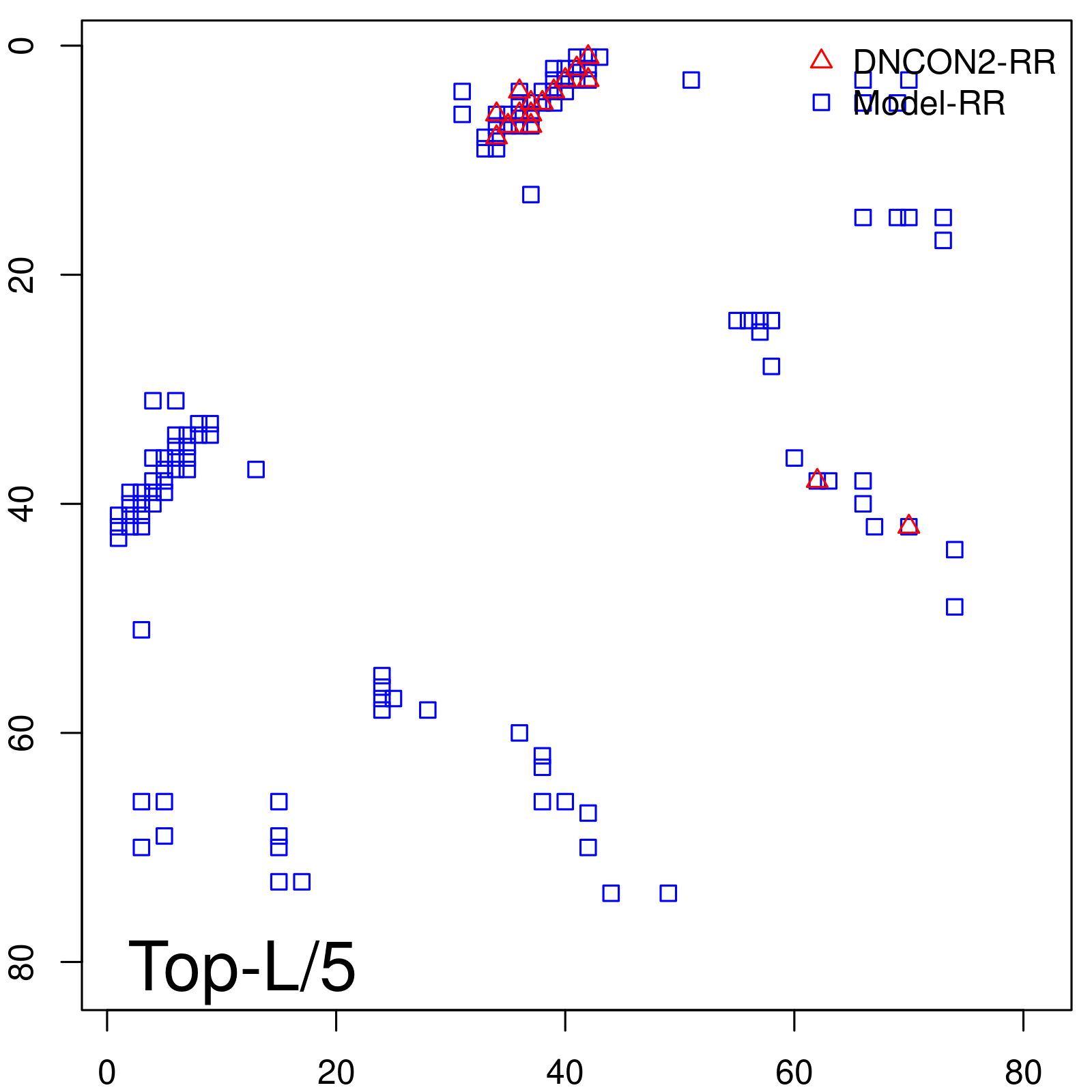
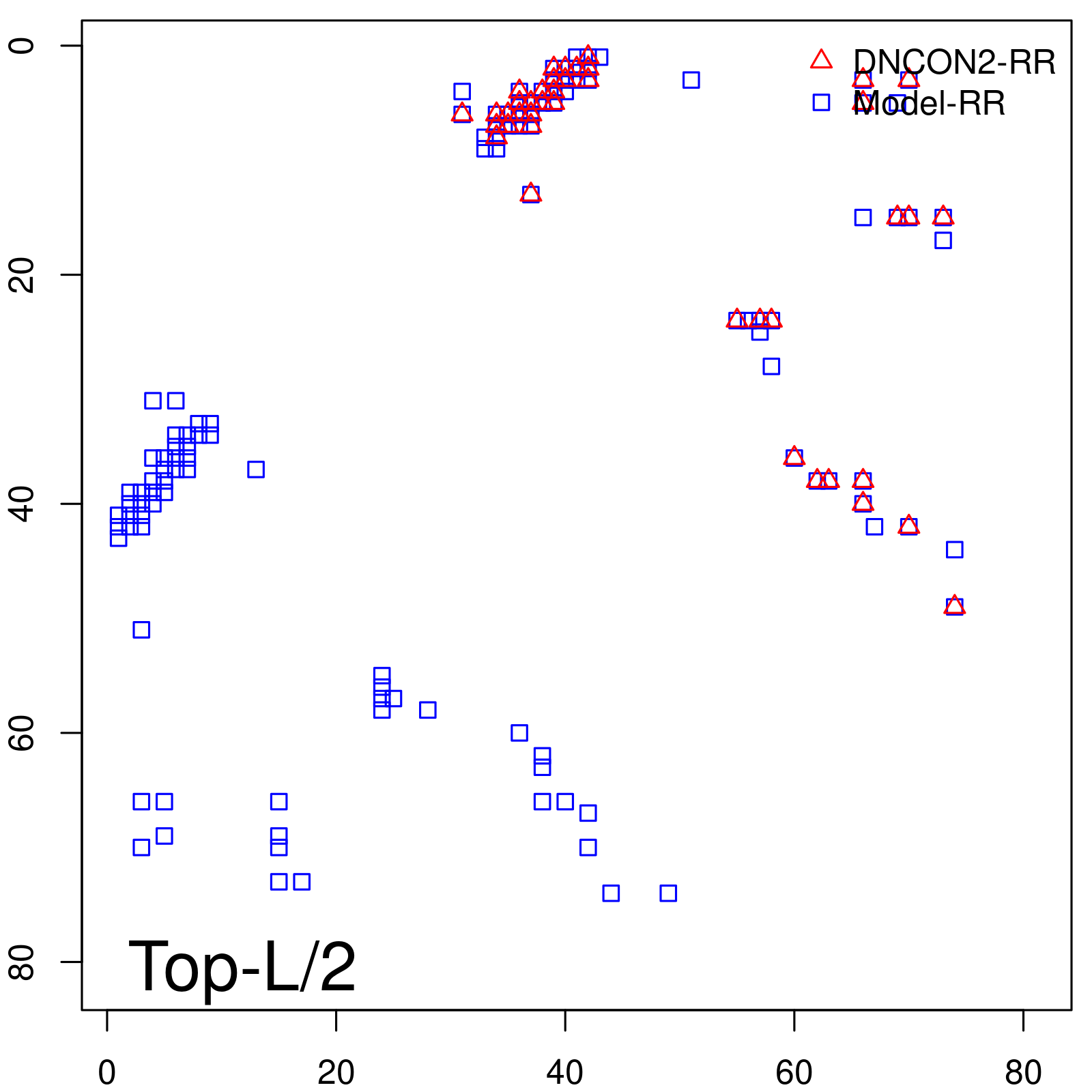
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 100.00 |
| TopL | 72.84 |
| Top2L | 36.42 |
( Alignment file  )
)
| Alignment | Number |
| N | 537 |
| Neff | 275 |
Model comparsion
| Model | TM-score |
| Model 2 | 0.9471 |
| Model 3 | 0.9523 |
| Model 4 | 0.9522 |
| Model 5 | 0.9581 |
| Average | 0.95242 |
Radius Gyration
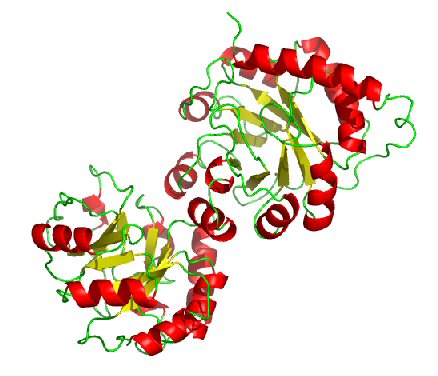
Similar pdbs
| PDB | TM-score |
| 2ozw | 0.63089 |
| 2ozx | 0.61549 |
| 2ai6 | 0.61488 |
| 2hw4 | 0.60752 |
| 2nmm | 0.60444 |
Predicted Top 2 Tertiary structure
|
|
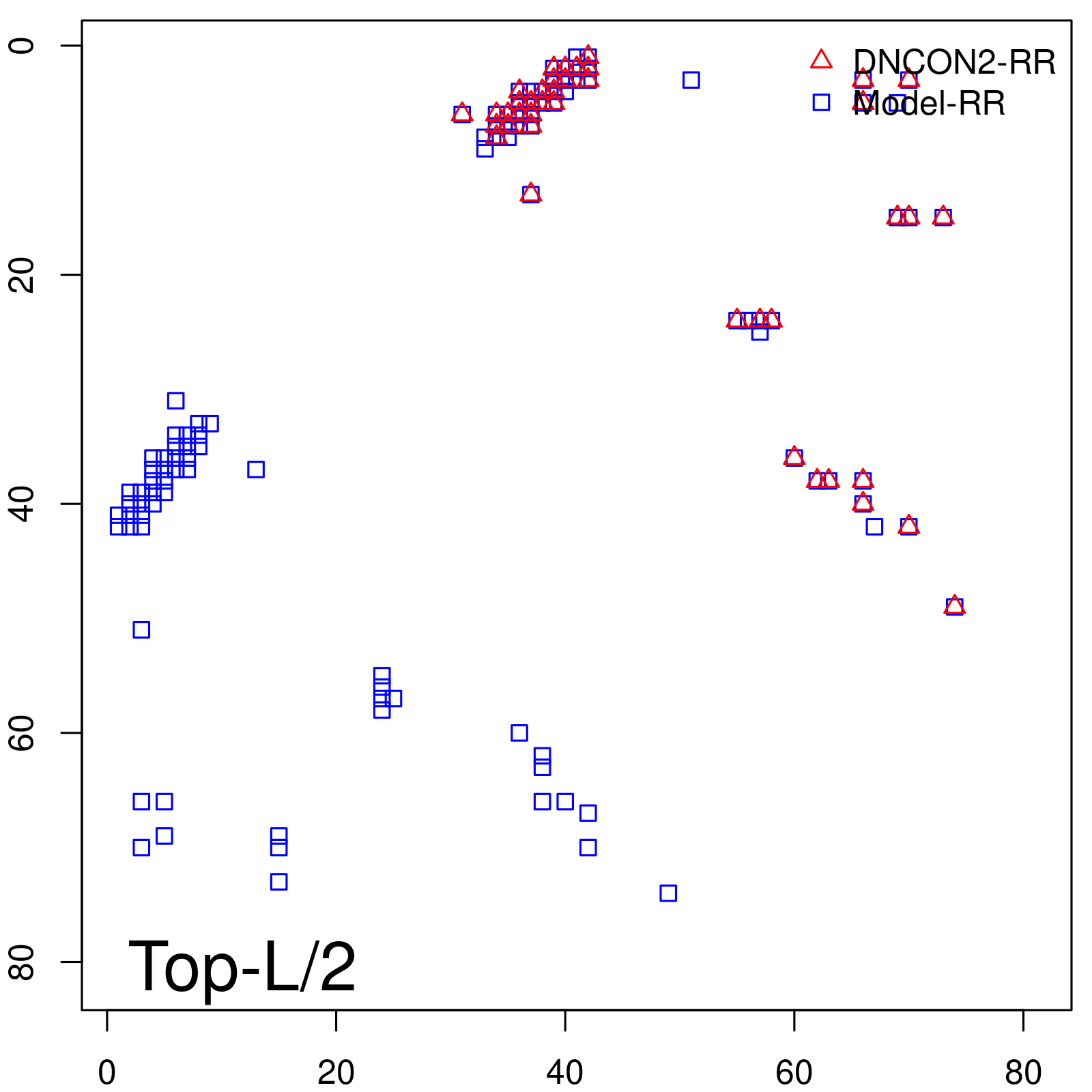
Predicted Contact Accuracy
|
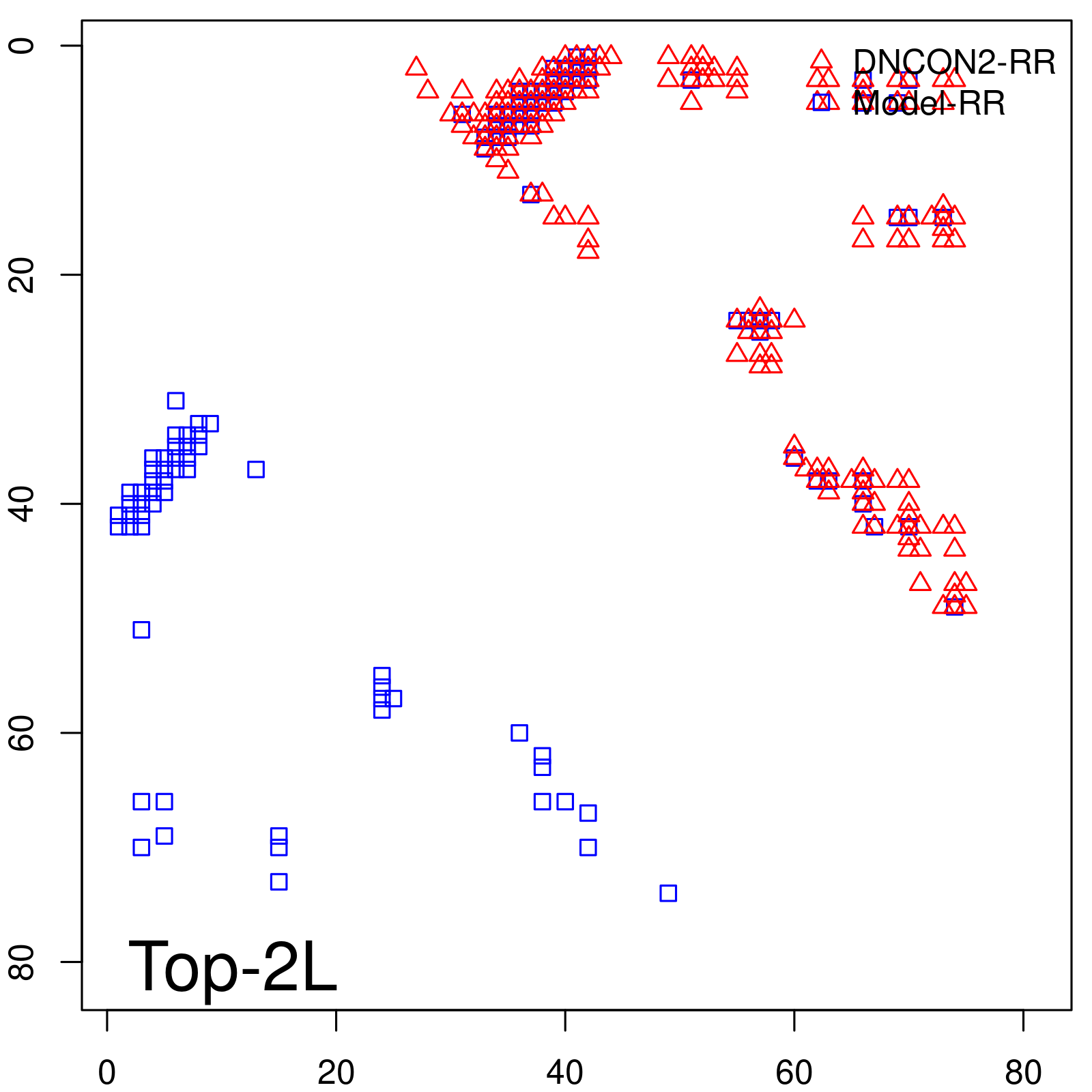
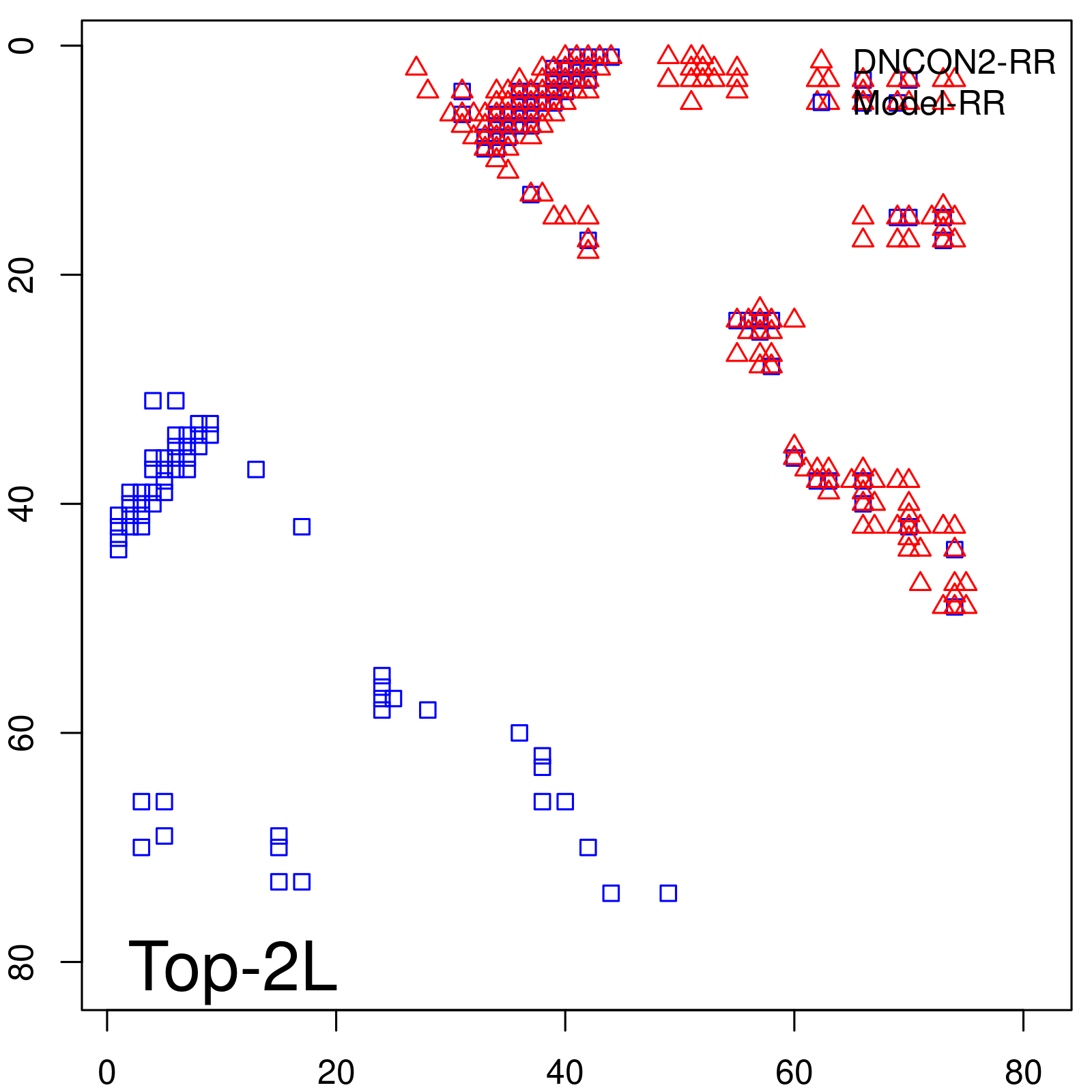
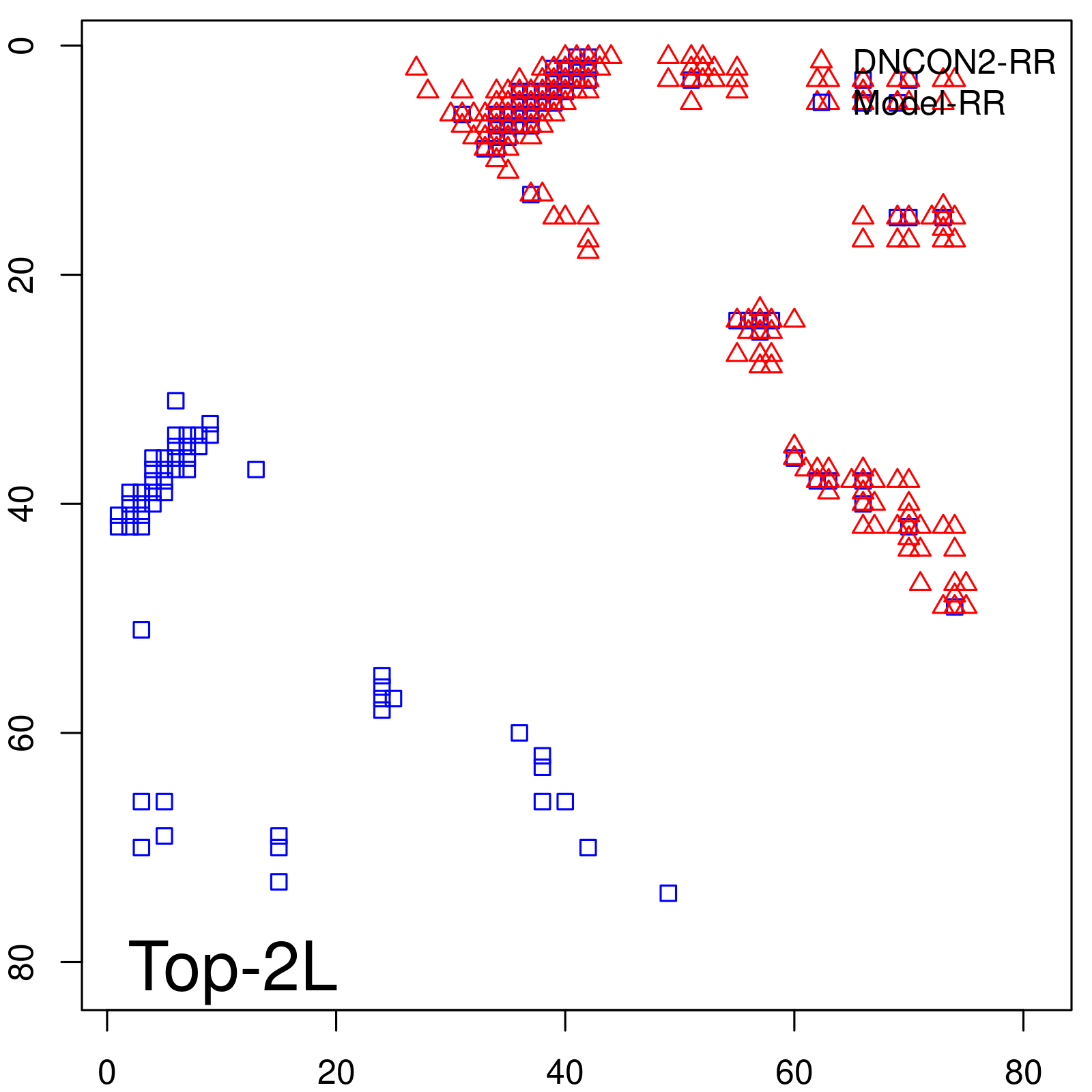
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 100.00 |
| TopL | 66.67 |
| Top2L | 33.33 |
( Alignment file  )
)
| Alignment | Number |
| N | 537 |
| Neff | 275 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9471 |
| Model 3 | 0.9469 |
| Model 4 | 0.9663 |
| Model 5 | 0.9475 |
| Average | 0.95195 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 2ozw | 0.62096 |
| 2ozx | 0.61776 |
| 2ai6 | 0.61126 |
| 2hw4 | 0.60053 |
| 2nmm | 0.59710 |
Predicted Top 3 Tertiary structure
|
|
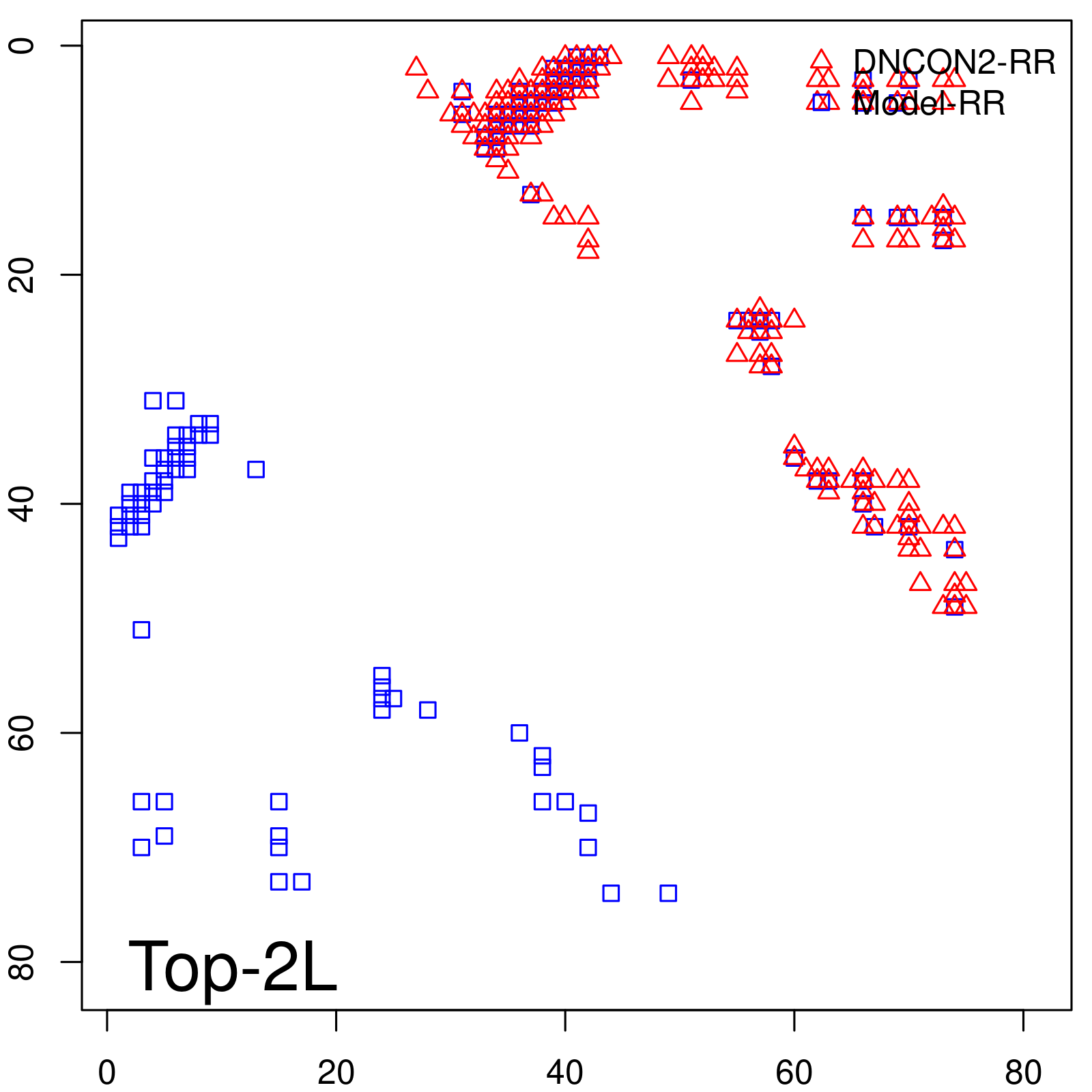
Predicted Contact Accuracy
|
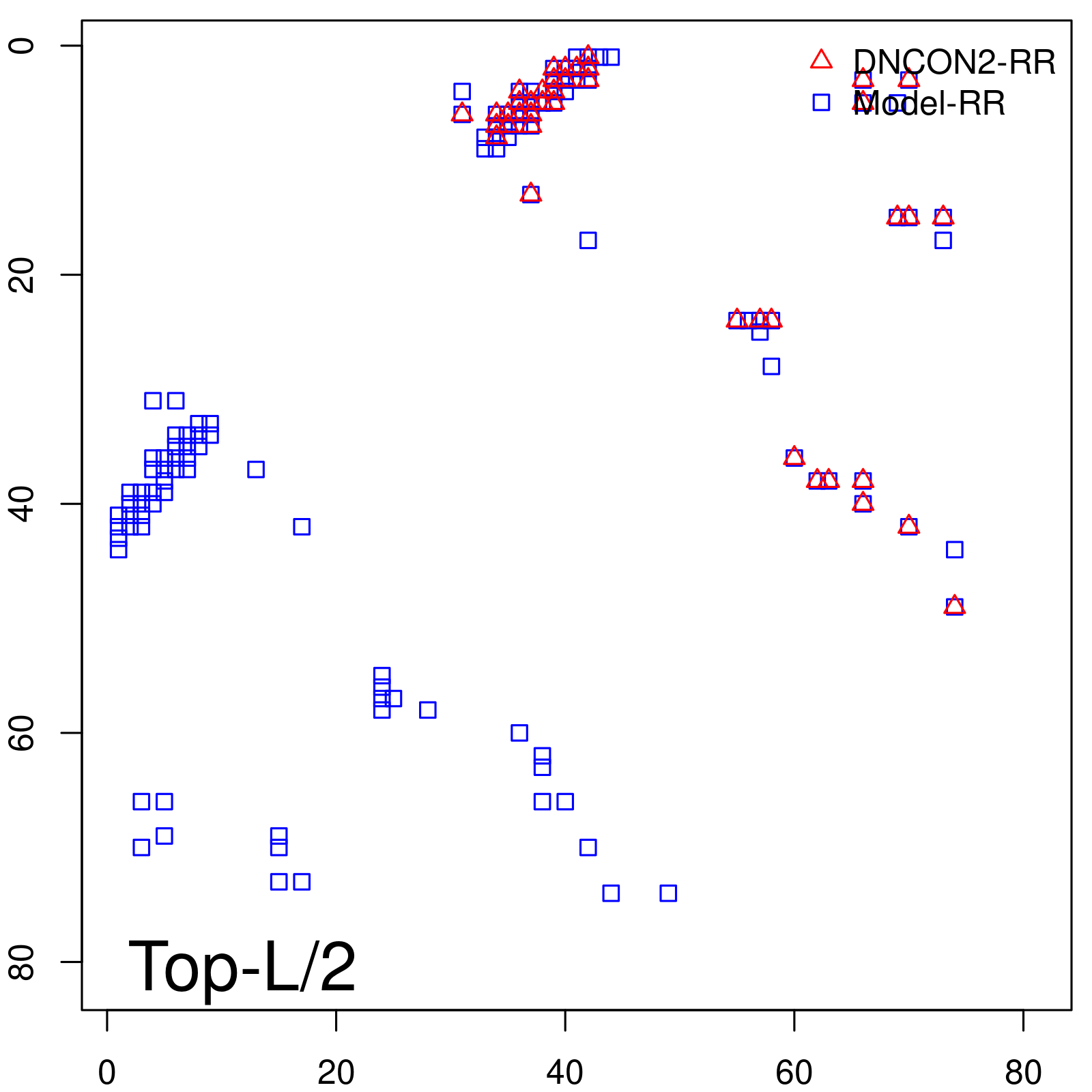
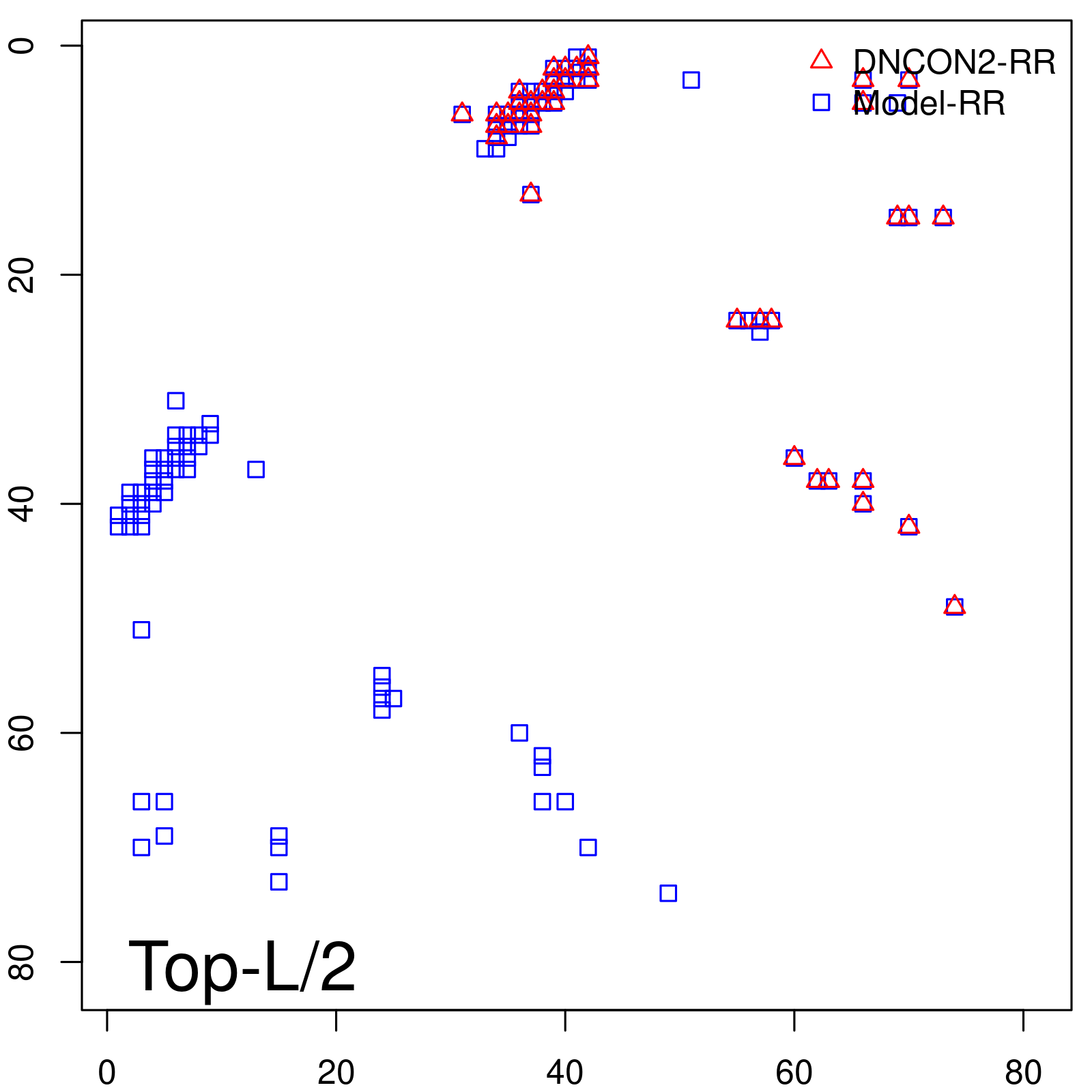
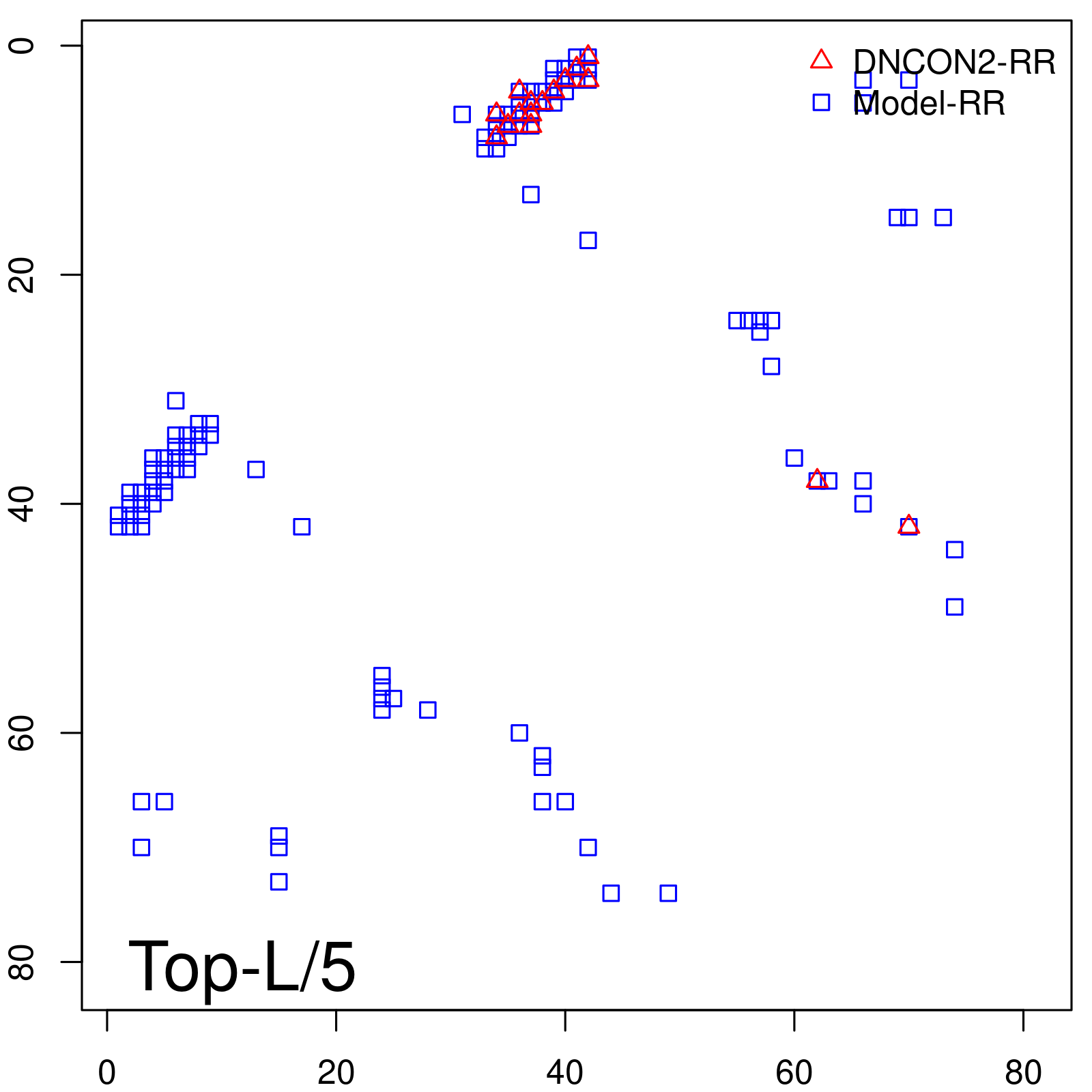
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 97.56 |
| TopL | 72.84 |
| Top2L | 36.42 |
( Alignment file  )
)
| Alignment | Number |
| N | 537 |
| Neff | 275 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9523 |
| Model 2 | 0.9469 |
| Model 4 | 0.9529 |
| Model 5 | 0.9576 |
| Average | 0.95243 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 2ozw | 0.61800 |
| 2ozx | 0.61157 |
| 2ai6 | 0.60713 |
| 2hw4 | 0.59603 |
| 2nmm | 0.59538 |
Predicted Top 4 Tertiary structure
|
|
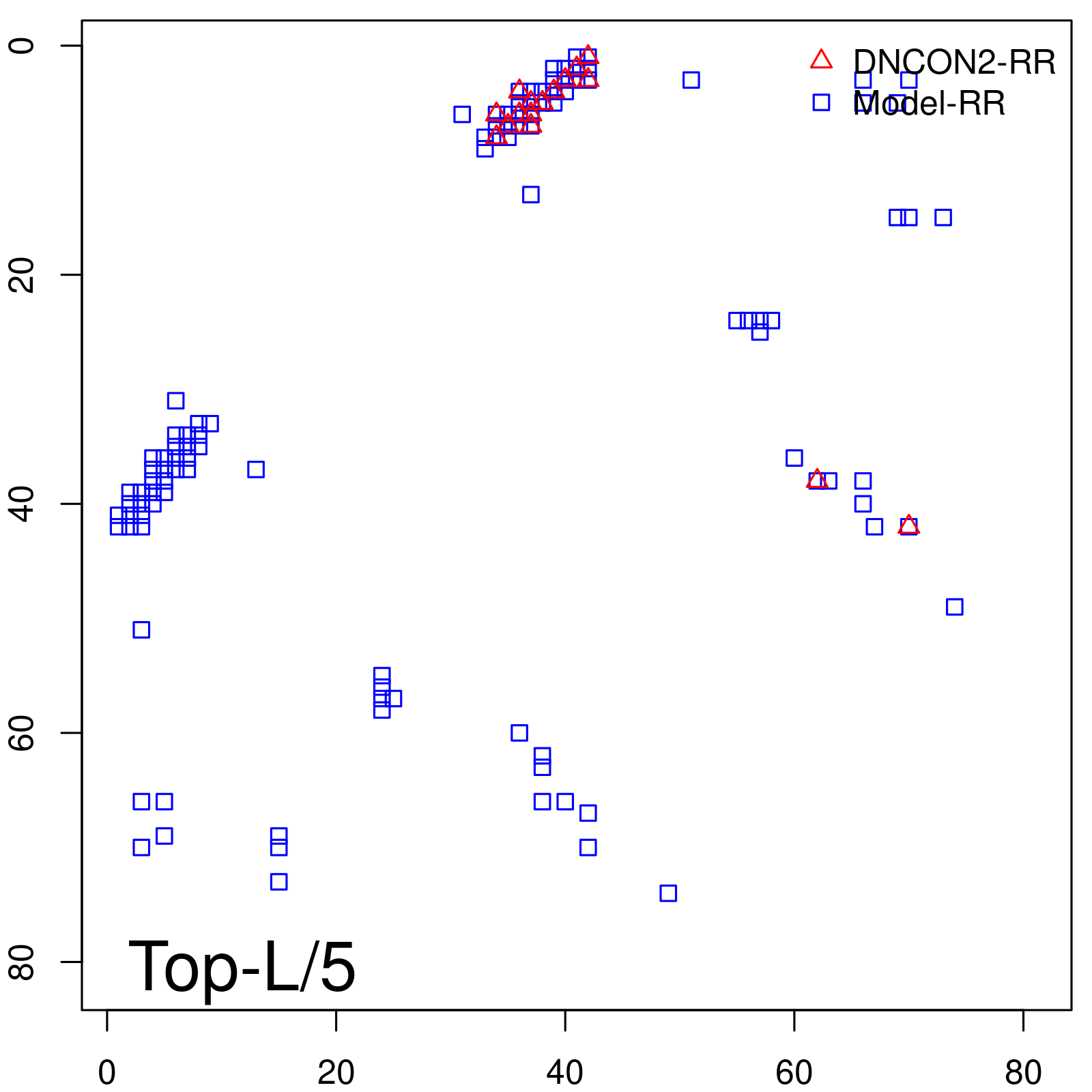
Predicted Contact Accuracy
|
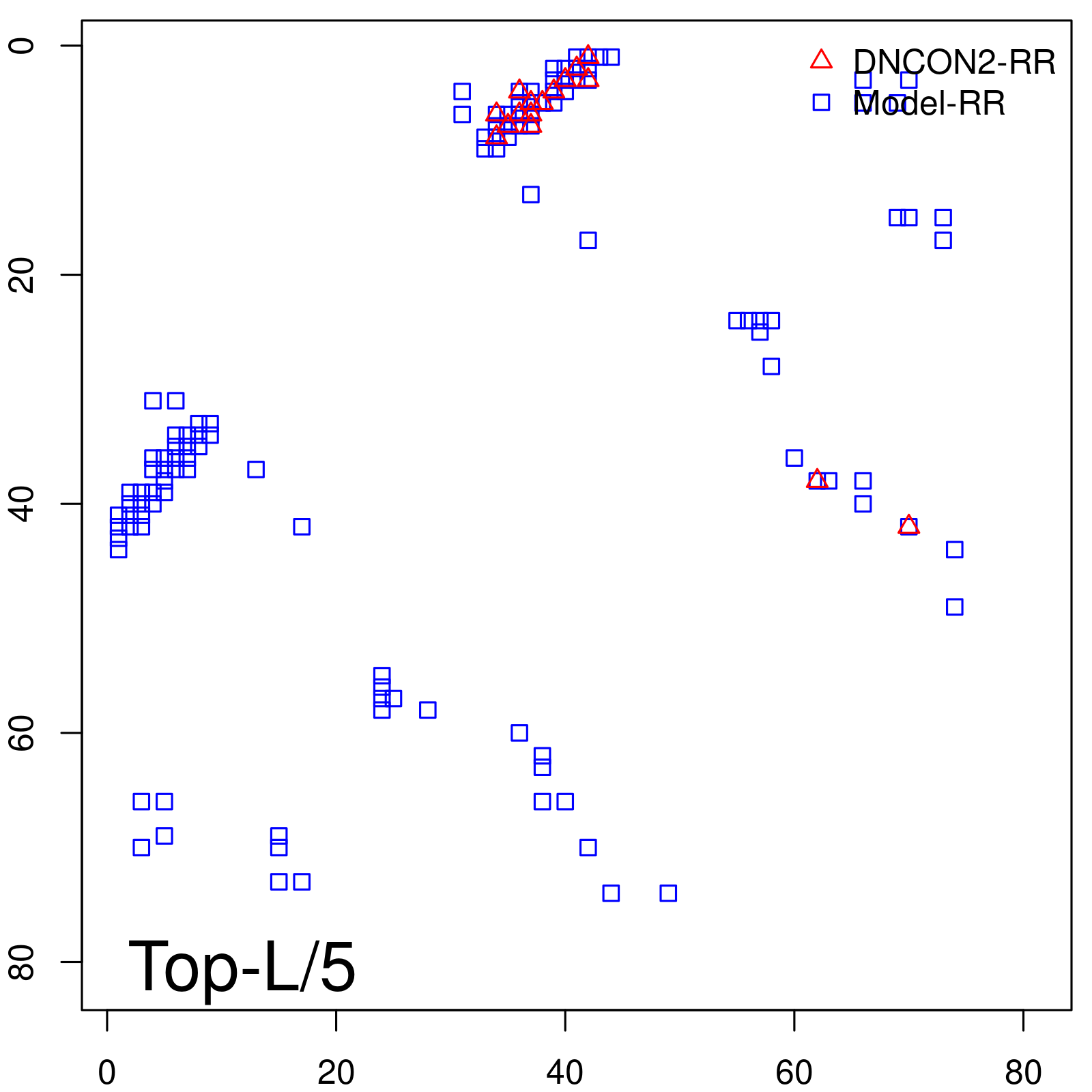
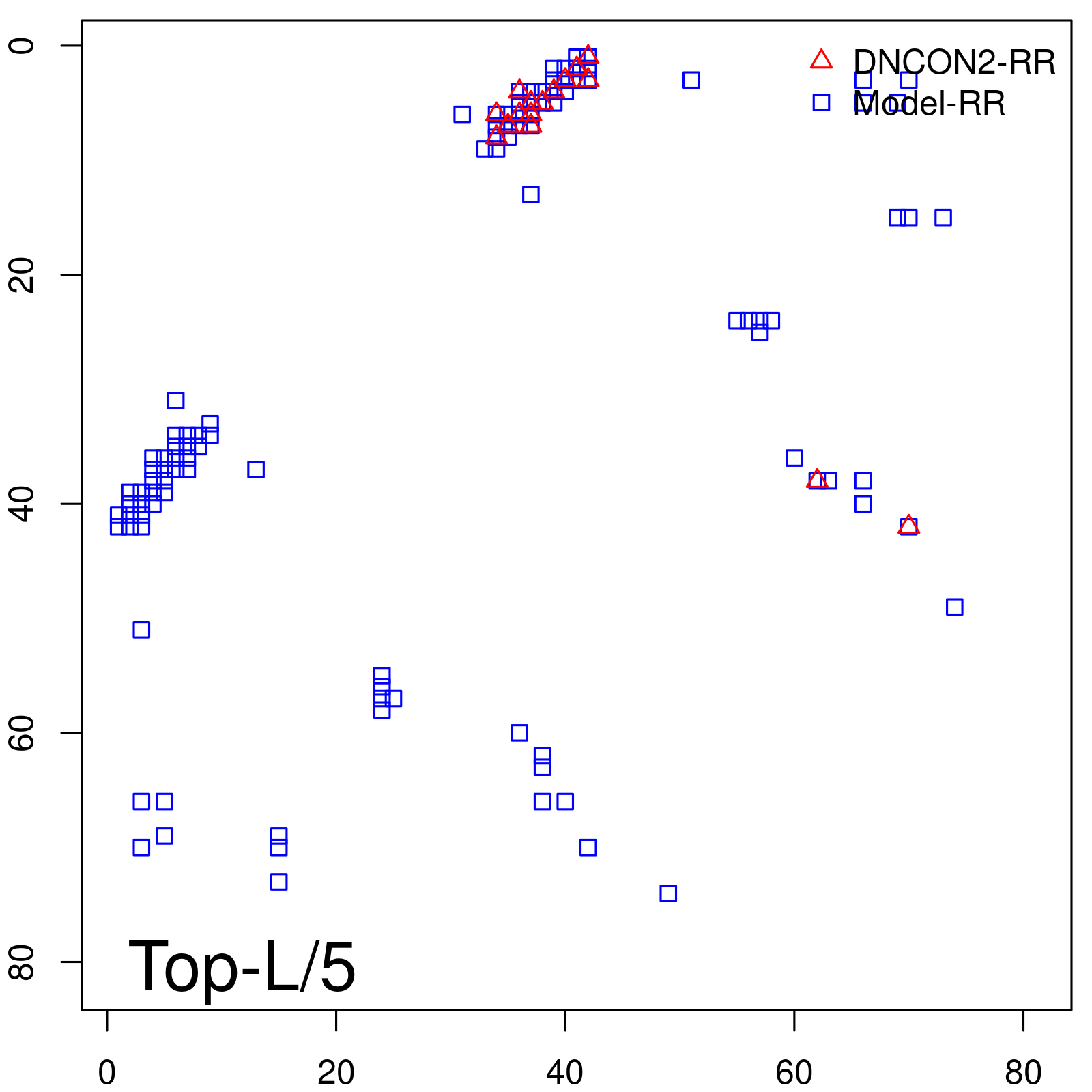
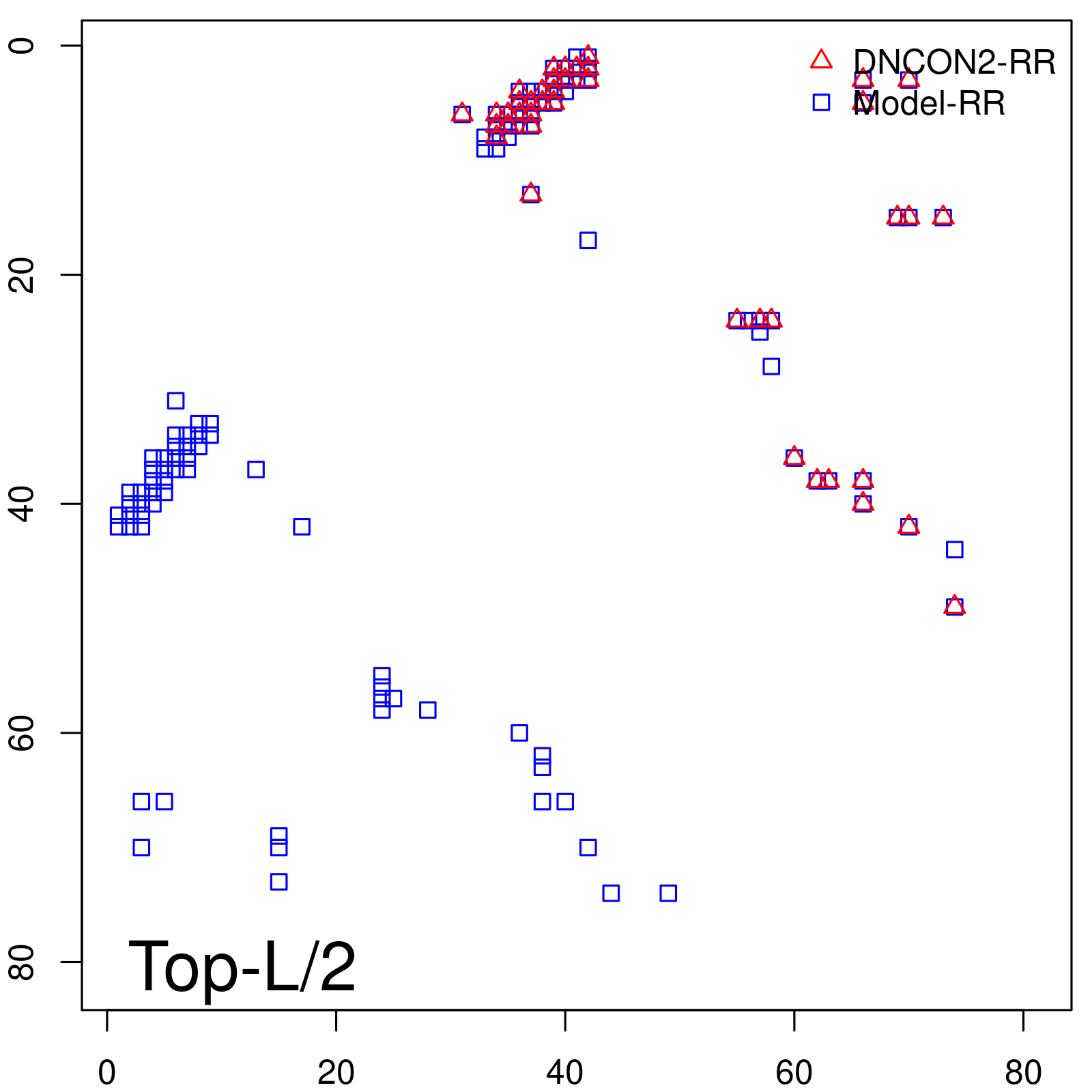
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 100.00 |
| TopL | 66.67 |
| Top2L | 33.33 |
( Alignment file  )
)
| Alignment | Number |
| N | 537 |
| Neff | 275 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9522 |
| Model 2 | 0.9663 |
| Model 3 | 0.9529 |
| Model 5 | 0.9546 |
| Average | 0.95650 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 2ozw | 0.62425 |
| 2ozx | 0.62117 |
| 2ai6 | 0.60819 |
| 2nmm | 0.60402 |
| 2hw4 | 0.60328 |
Predicted Top 5 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 100.00 |
| TopL/2 | 100.00 |
| TopL | 67.90 |
| Top2L | 33.95 |
( Alignment file  )
)
| Alignment | Number |
| N | 537 |
| Neff | 275 |
Model comparsion
| Model | TM-score |
| Model 1 | 0.9581 |
| Model 2 | 0.9475 |
| Model 3 | 0.9576 |
| Model 4 | 0.9546 |
| Average | 0.95445 |
Radius Gyration
Similar pdbs
| PDB | TM-score |
| 2ozw | 0.61914 |
| 2ozx | 0.61527 |
| 2ai6 | 0.60750 |
| 2nmm | 0.59690 |
| 2hw4 | 0.59471 |