Results of Structure Prediction
Domain Boundary prediction ( View  )
)

Protein sequence
| 1-60: | M | D | A | T | S | I | I | T | Q | V | S | R | D | D | E | Q | L | N | V | Y | P | S | Y | P | N | D | K | D | D | V | D | R | L | K | P | Q | K | R | G | L | F | H | S | L | L | C | C | W | R | R | N | R | T | K | T | N | Q | N | G | T |
| 61-119: | Q | I | D | G | S | T | T | P | P | P | L | P | D | Q | Q | R | Y | L | L | P | Q | V | R | L | T | D | M | H | R | K | C | M | V | I | D | L | D | E | T | L | V | H | S | S | F | K | P | I | P | N | A | D | F | I | V | P | V | E | I | D |
| 121-179: | G | T | V | H | Q | V | Y | V | L | K | R | P | H | V | D | E | F | L | Q | K | M | G | E | L | Y | E | C | V | L | F | T | A | S | L | A | K | Y | A | D | P | V | A | D | L | L | D | K | W | N | V | F | R | A | R | L | F | R | E | S | C |
| 181-239: | V | Y | Y | R | G | N | Y | I | K | D | L | N | R | L | G | R | D | L | Q | K | I | V | I | V | D | N | S | P | A | S | Y | I | F | H | P | D | N | A | V | P | V | K | S | W | F | D | D | V | T | D | C | E | L | R | E | L | I | P | L | F |
| 241-299: | E | K | L | S | K | V | D | S | V | Y | S | V | L | C | N | S | N | Q | P | L | N | N | Q | T | N | Q | Q | Q | H | P | Q | E | L | Q | Q | A | P | N | Q | L | H | Q | Q | L | Q | Q | Q | Q | Q | Q | Q | T | I | S | A | T | T | V | I | T |
| 301-329: | Q | A | T | T | L | S | A | P | T | M | L | N | Q | Q | Q | T | S | P | P | S | P | Q | S | E | L | L | Q | K | T |
Secondary structure prediction (H: Helix E: Strand C: Coil)
| 1-60: | C | C | C | C | C | E | E | E | E | E | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | H | H | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H | C | C | C | C | C | H | C | C | C | C | C | C |
| 61-119: | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | E | E | E | E | C | C | E | C | C | C | E | E | E | E | C | C | C | C | C | C | C | C | E | E | E | E | E | E | E | C |
| 121-179: | C | E | E | E | E | E | E | E | E | E | C | C | C | H | H | H | H | H | H | H | H | H | H | H | C | E | E | E | E | E | C | C | C | C | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | E | E | E | E | E | C | H | H | H | C |
| 181-239: | E | E | E | C | C | E | E | E | C | C | H | H | H | C | C | C | C | H | H | H | E | E | E | E | E | C | C | H | H | H | H | C | C | C | H | H | H | E | E | E | C | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | H | H | H | H |
| 241-299: | H | H | H | H | C | C | C | C | C | H | H | H | H | H | H | H | C | C | C | H | H | H | H | H | H | C | C | C | C | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | C | C | C | C | C | C | C | C | C | C |
| 301-329: | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | H | H | H | H | H | C | C | C |
Solvent accessibility prediction (e: Exposed b: Buried)
| 1-60: | E | E | E | E | B | B | B | B | B | B | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | E | B | B | B | B | B | B | B | B | B | B | E | E | E | E | E | E | E | E | E | E | E |
| 61-119: | E | B | E | E | E | E | E | E | E | E | B | E | E | E | E | E | E | B | B | E | E | B | E | E | E | B | E | E | E | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | E | E | E | E | E | E | B | E | B | E | B | E | B | E | E | E |
| 121-179: | E | E | E | E | E | B | B | B | B | B | B | B | E | B | E | E | B | B | E | E | B | B | E | B | B | B | B | B | B | B | B | B | B | E | E | E | B | B | E | E | B | B | E | E | B | B | E | E | E | B | B | E | B | E | B | B | E | E | B | B |
| 181-239: | E | E | E | E | E | E | E | B | B | B | B | E | E | B | E | B | E | E | E | B | B | B | B | B | B | B | E | E | E | B | B | E | E | B | E | E | B | B | B | E | B | E | E | B | E | E | E | E | E | B | E | E | B | E | E | B | B | E | E | B |
| 241-299: | E | E | B | B | E | B | E | E | B | B | E | E | B | E | E | B | E | E | E | E | E | E | E | E | E | E | E | E | B | E | E | E | B | E | E | E | E | E | E | B | E | E | E | B | E | E | E | E | E | E | E | E | E | E | E | E | E | E | B | E |
| 301-329: | E | B | B | E | B | E | B | B | E | E | B | E | E | E | E | E | E | E | E | E | E | E | E | E | B | E | E | E | E |
Disorder prediction (N: Normal T: Disorder)
| 1-60: | T | T | T | N | N | N | N | N | N | N | N | N | N | T | N | T | N | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T | T | T | T | T | T |
| 61-119: | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 121-179: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 181-239: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 241-299: | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | N | N | N | N | N | N | N | N | N | N | N | N | N |
| 301-329: | N | N | N | N | N | N | N | N | N | N | N | N | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
Select domain
Predicted Top 1 Tertiary structure
|
|
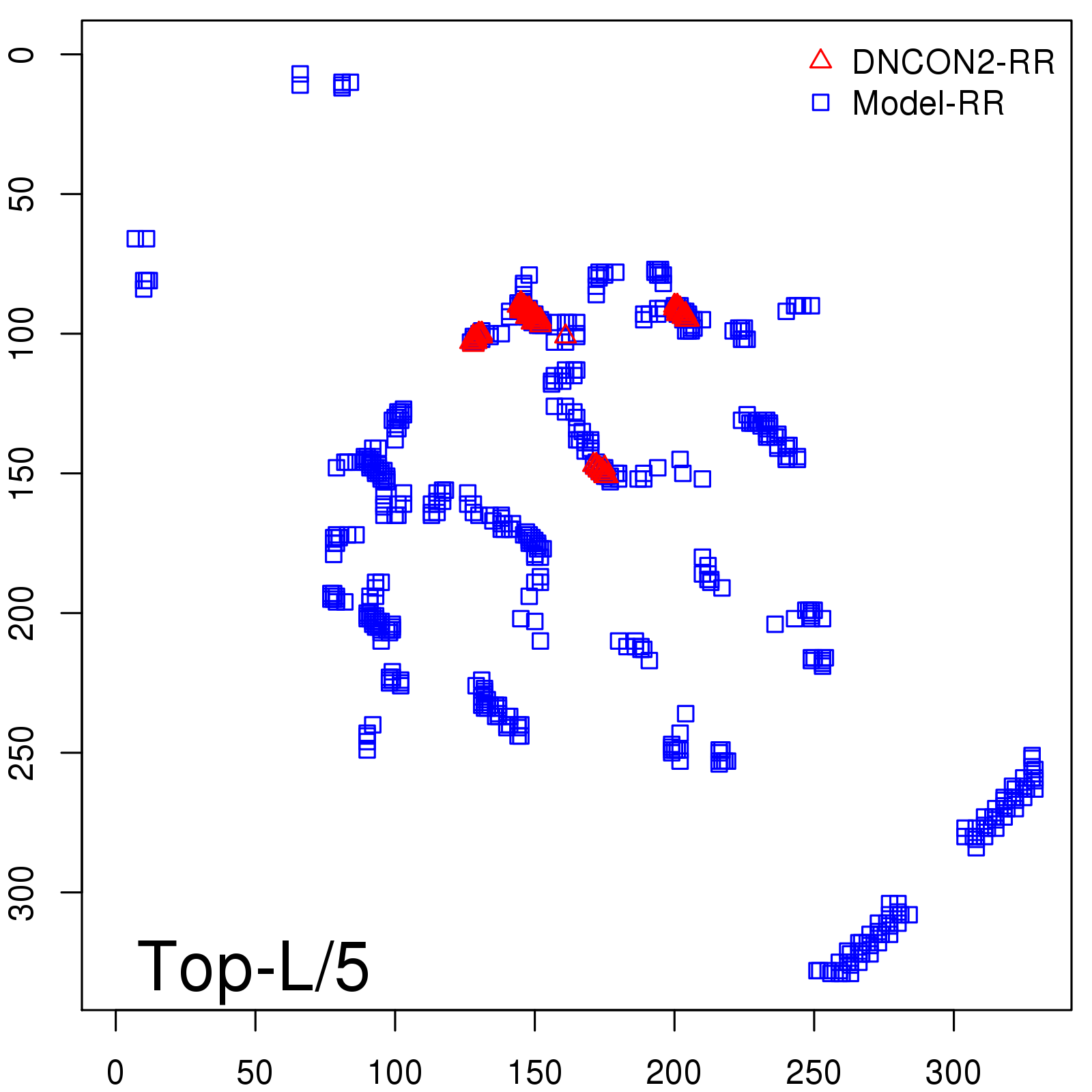
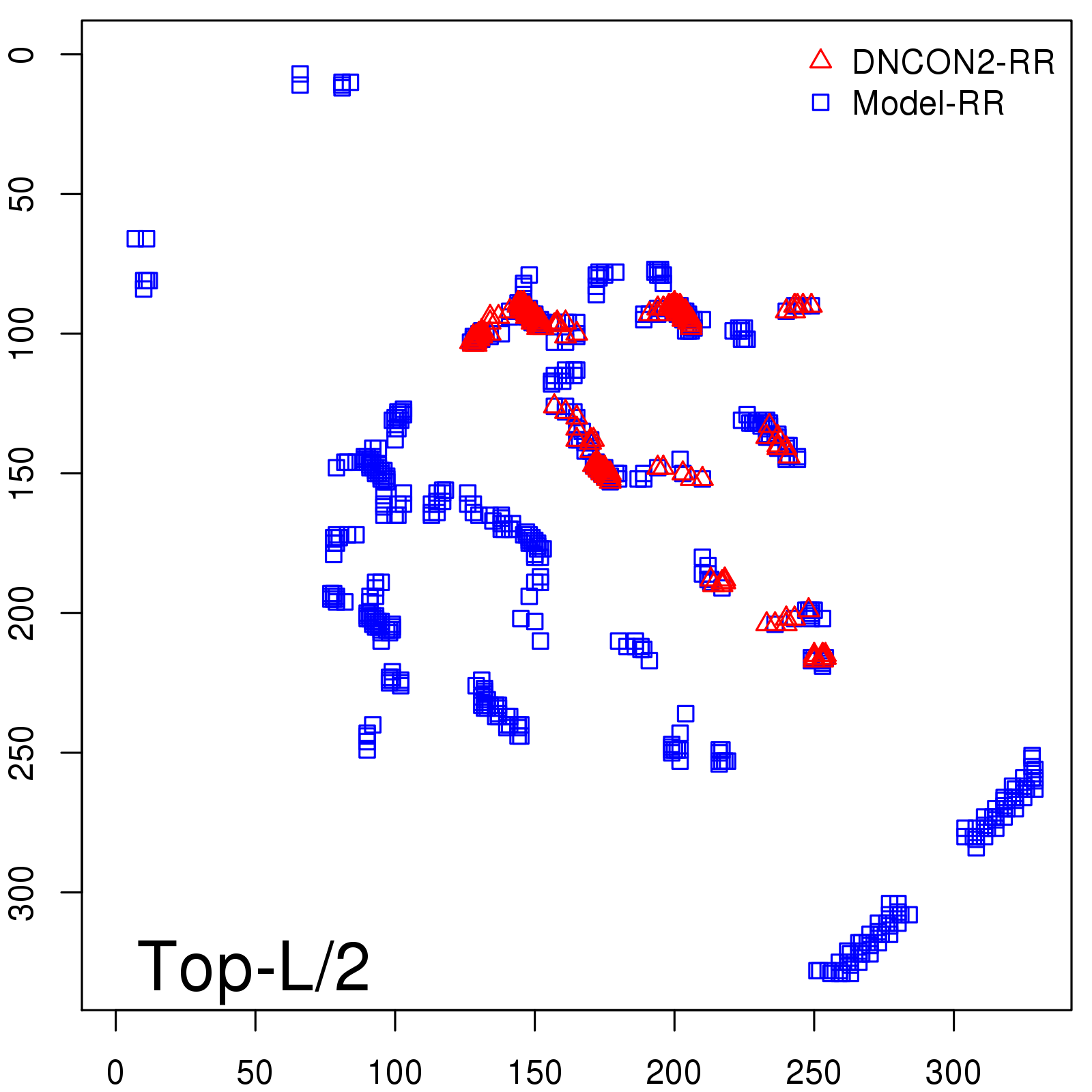
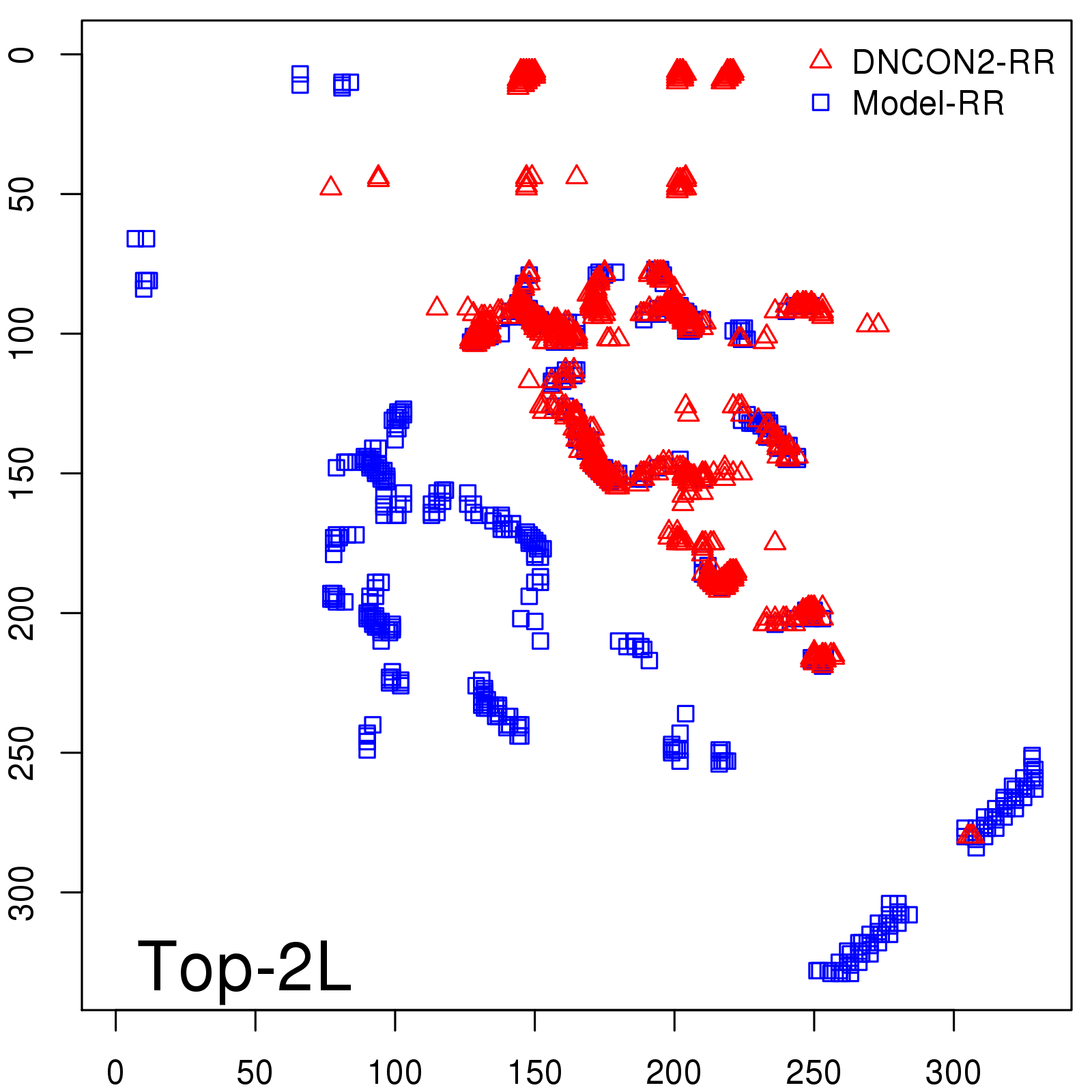
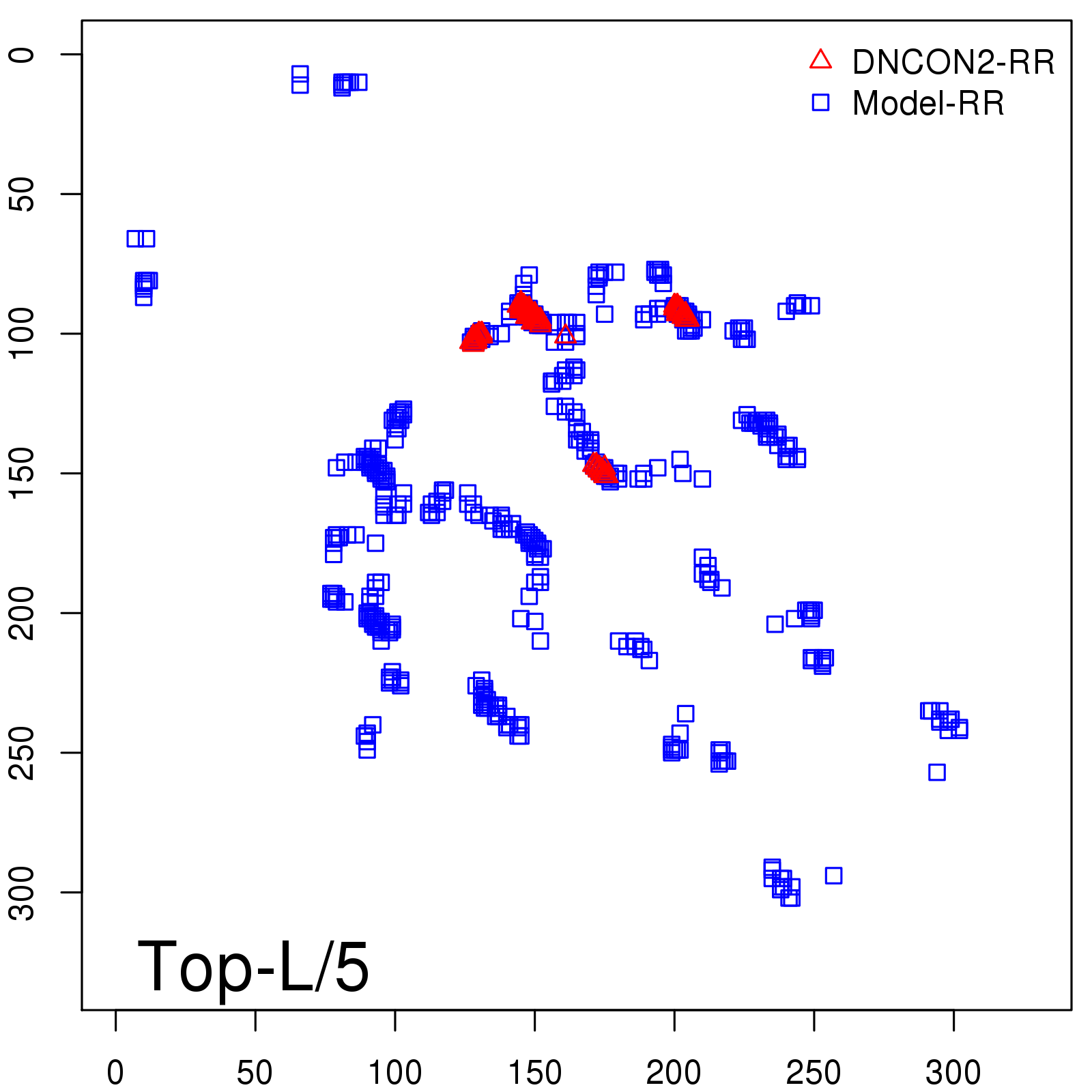
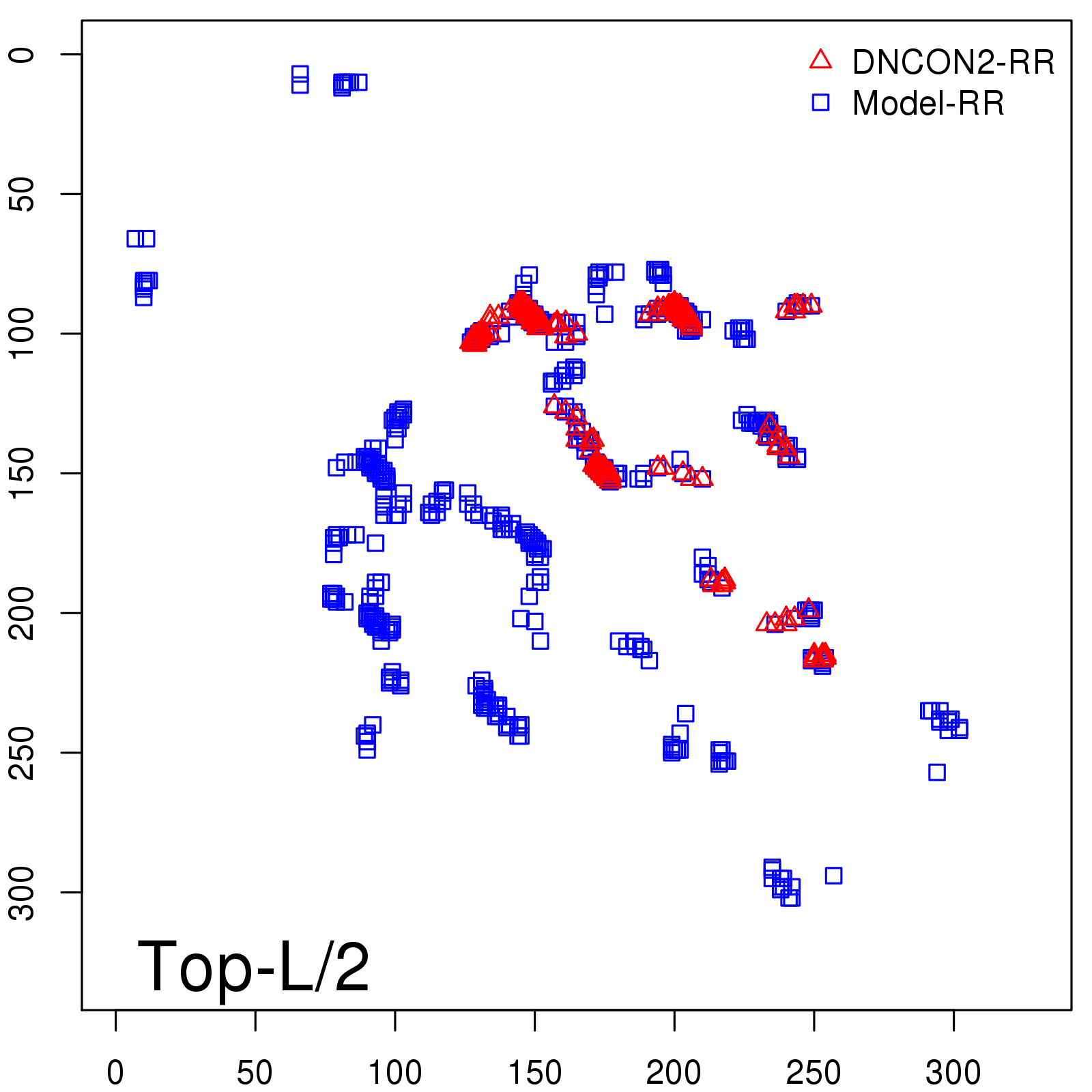
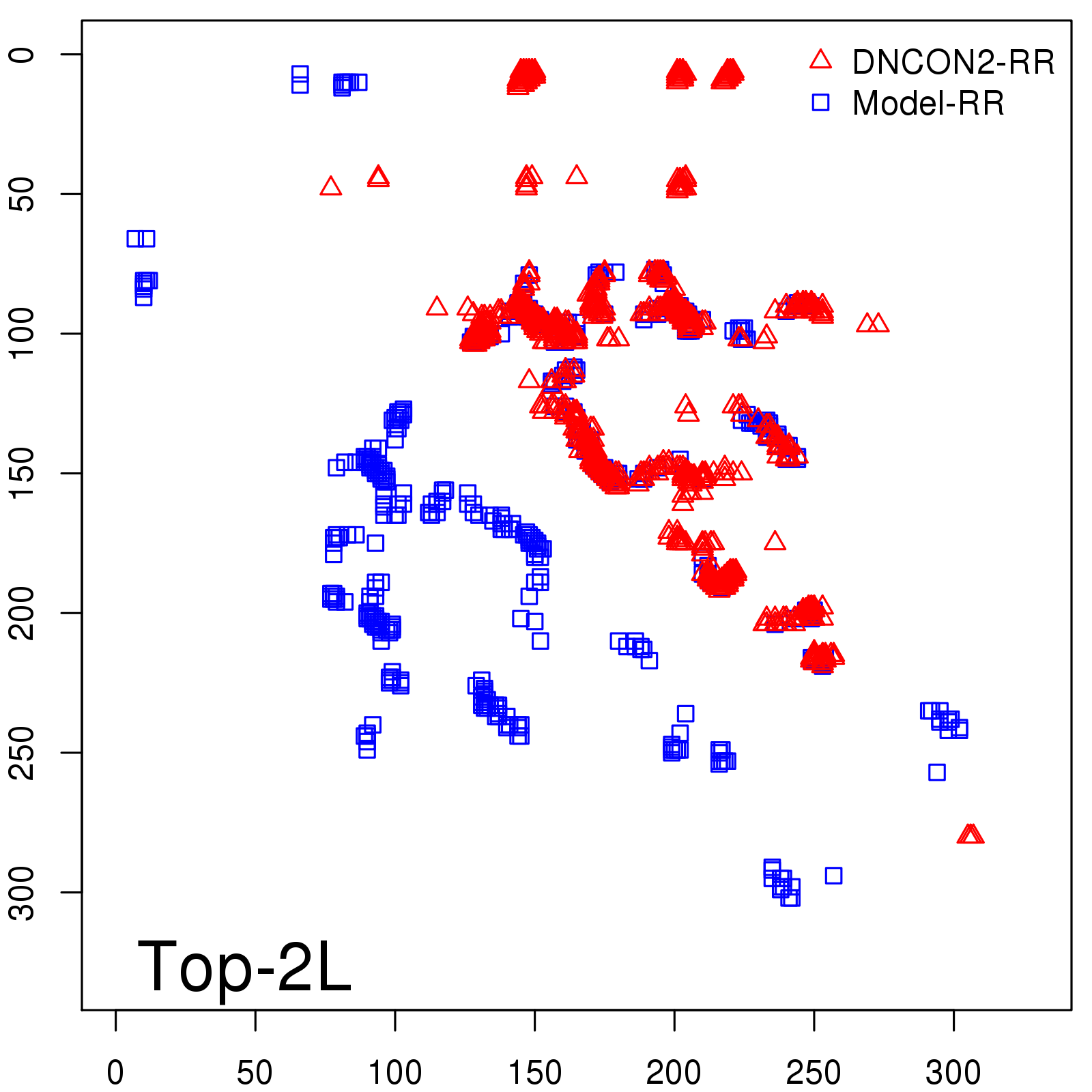
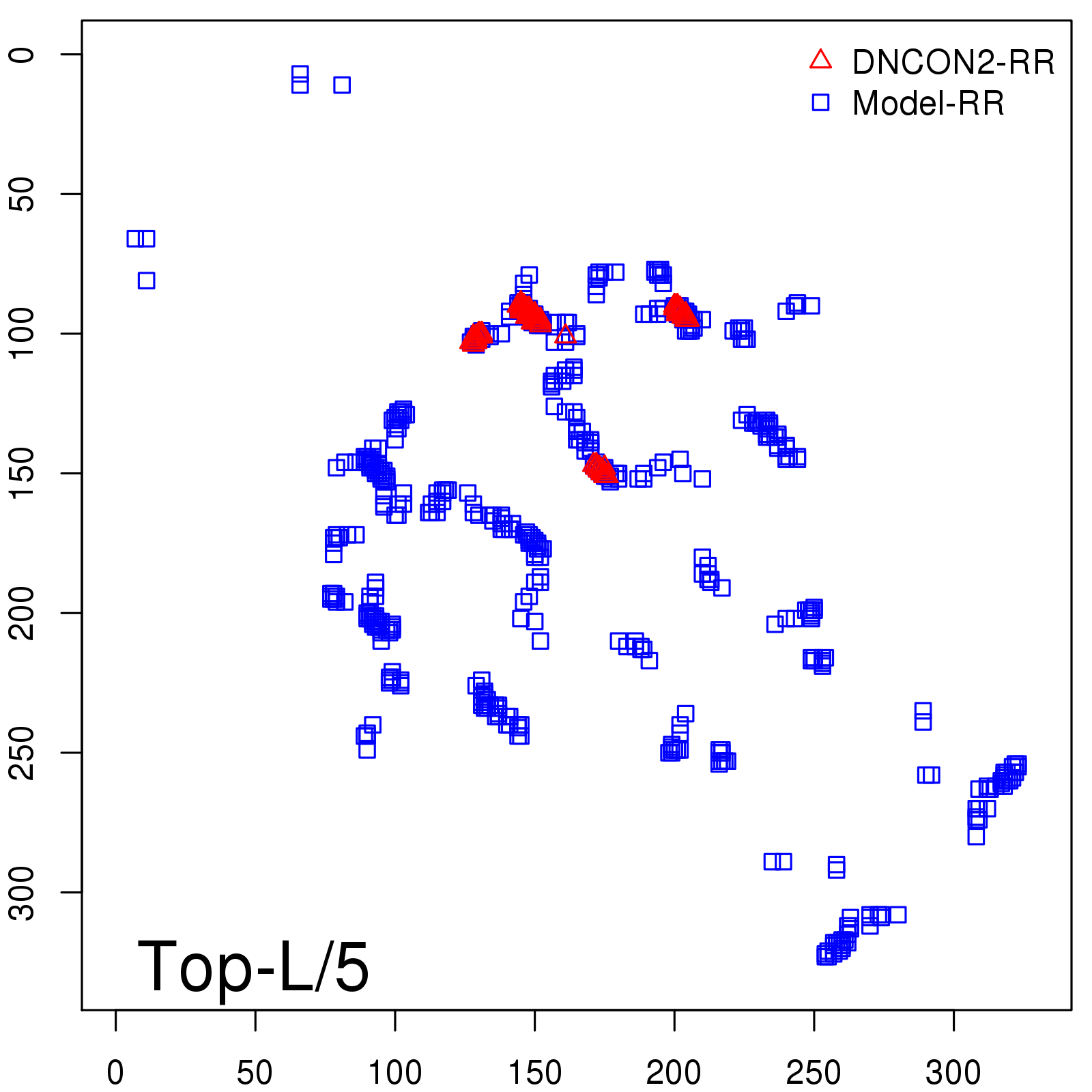
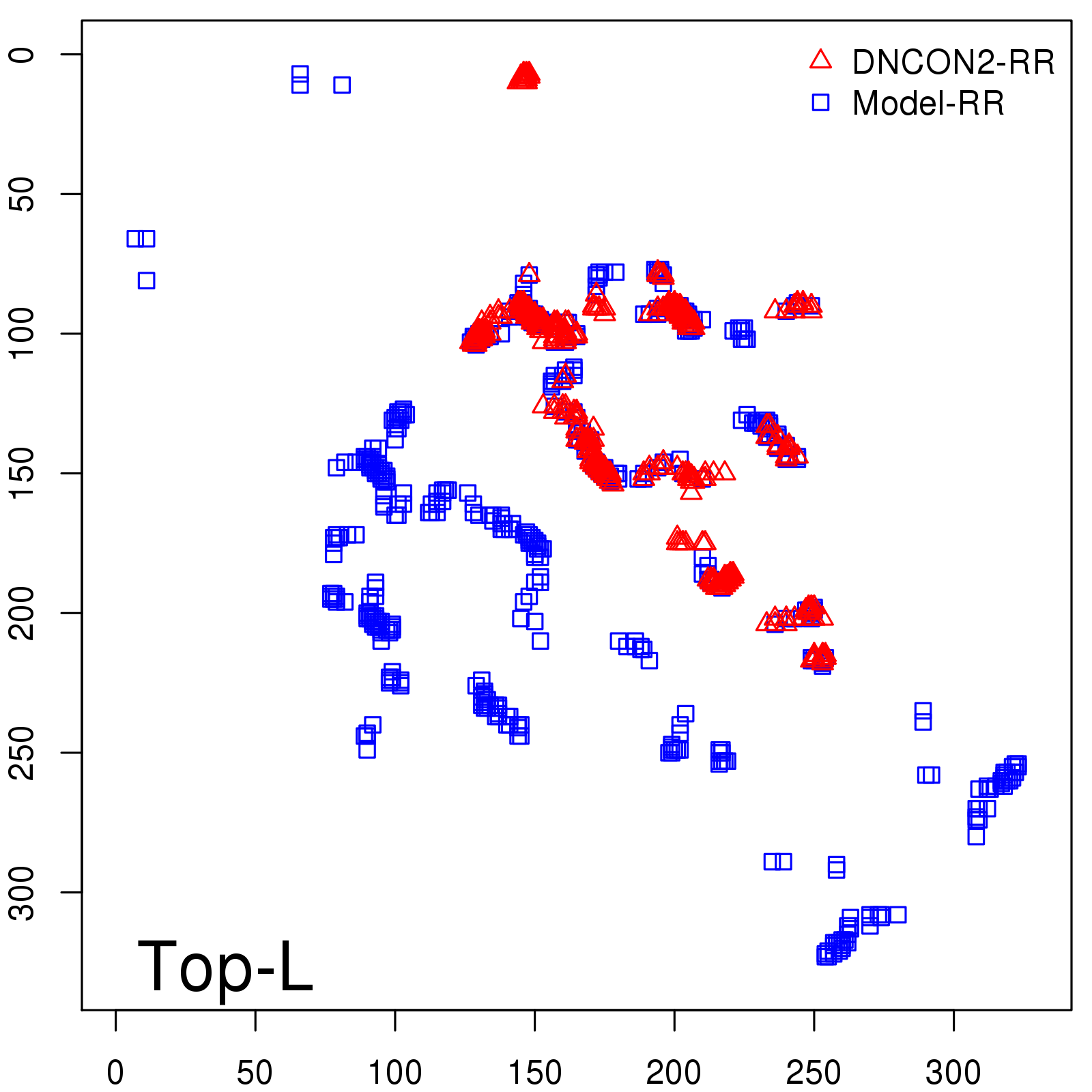
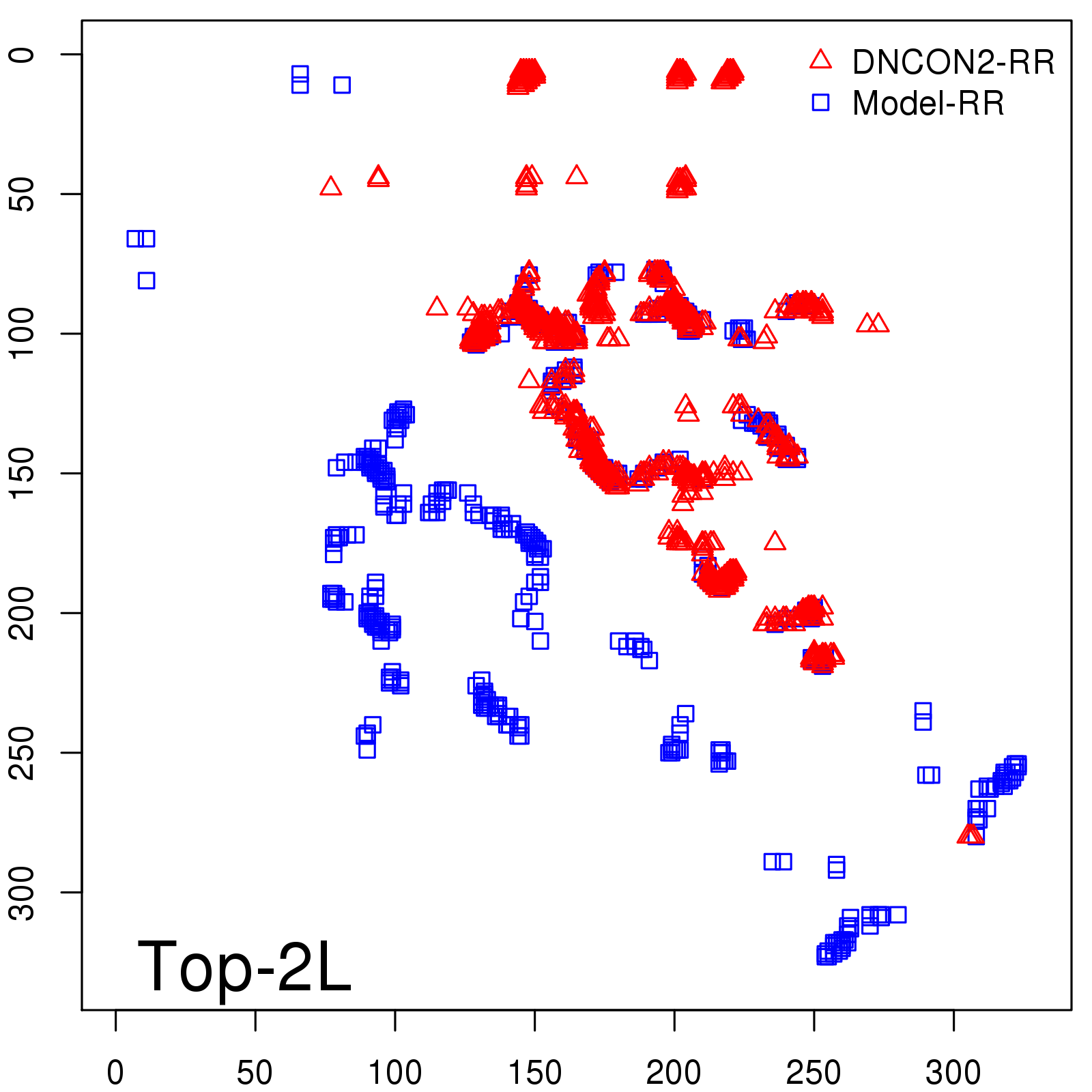
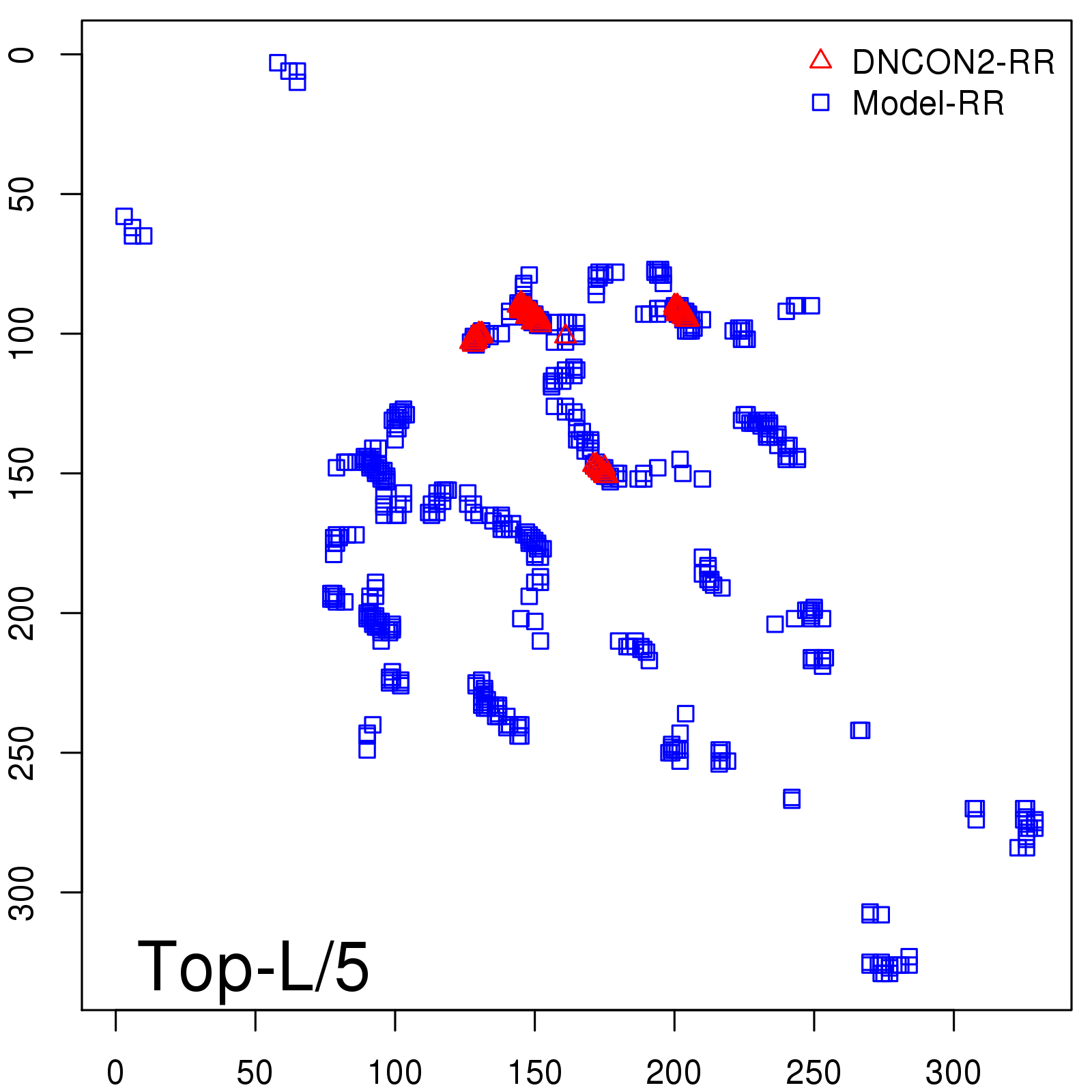
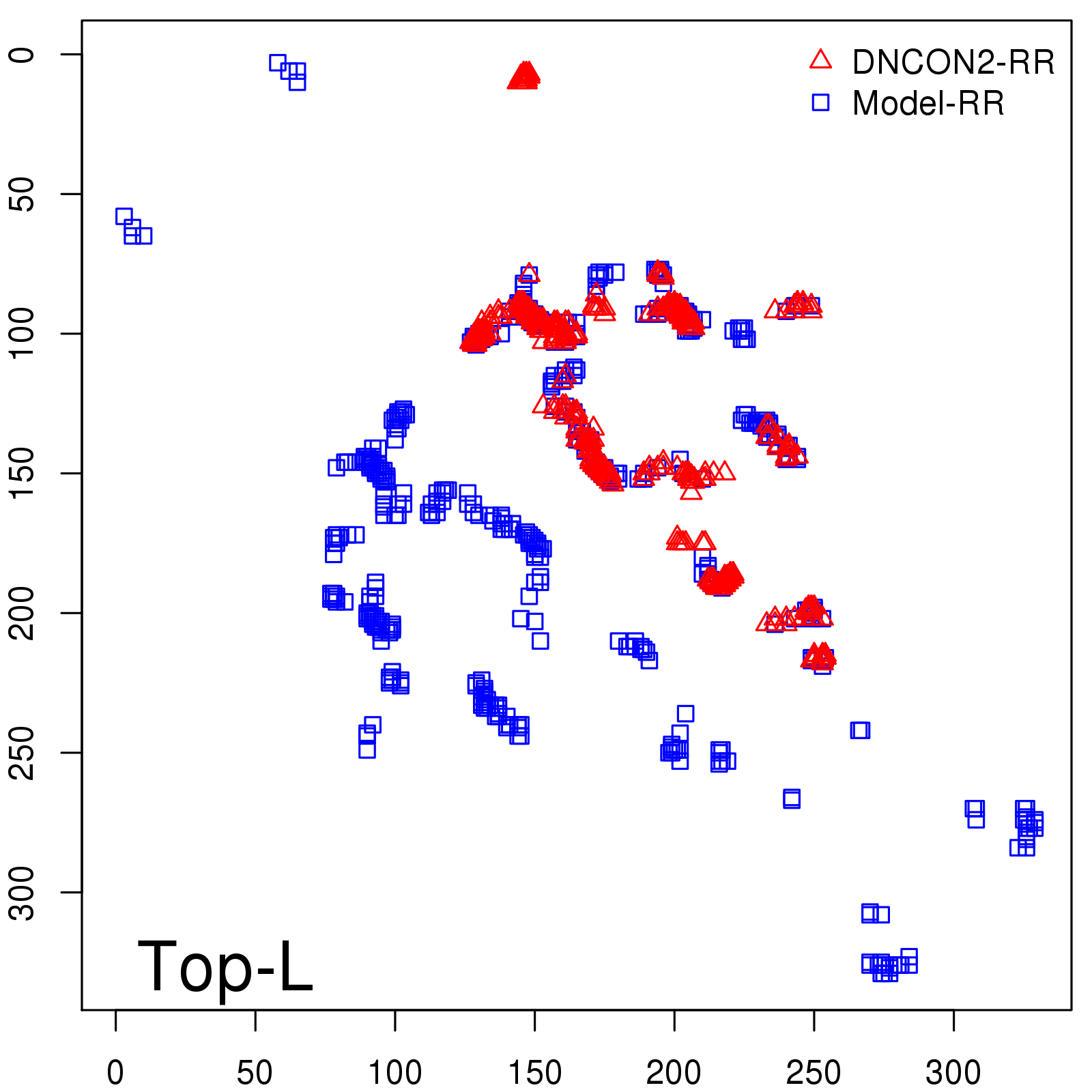
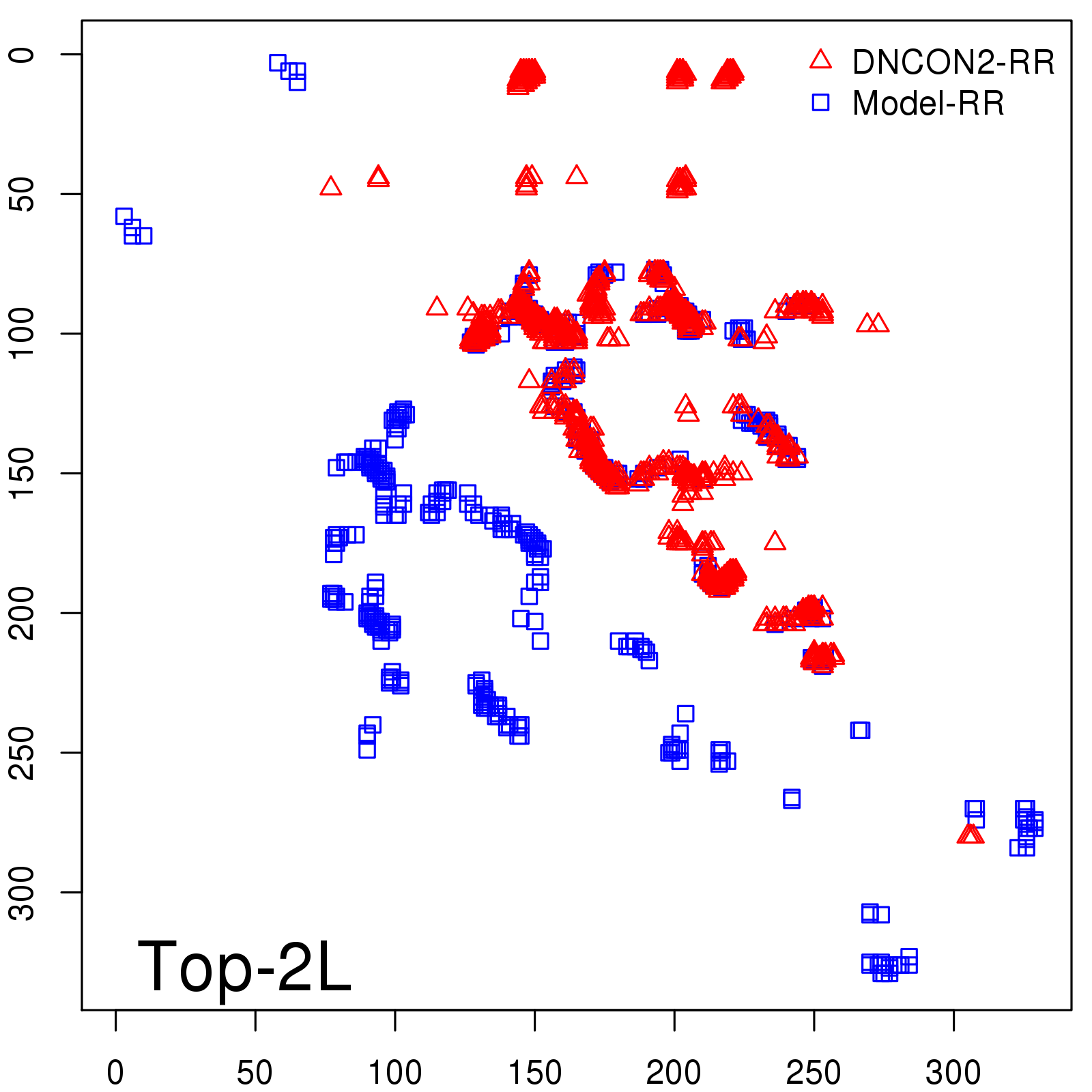
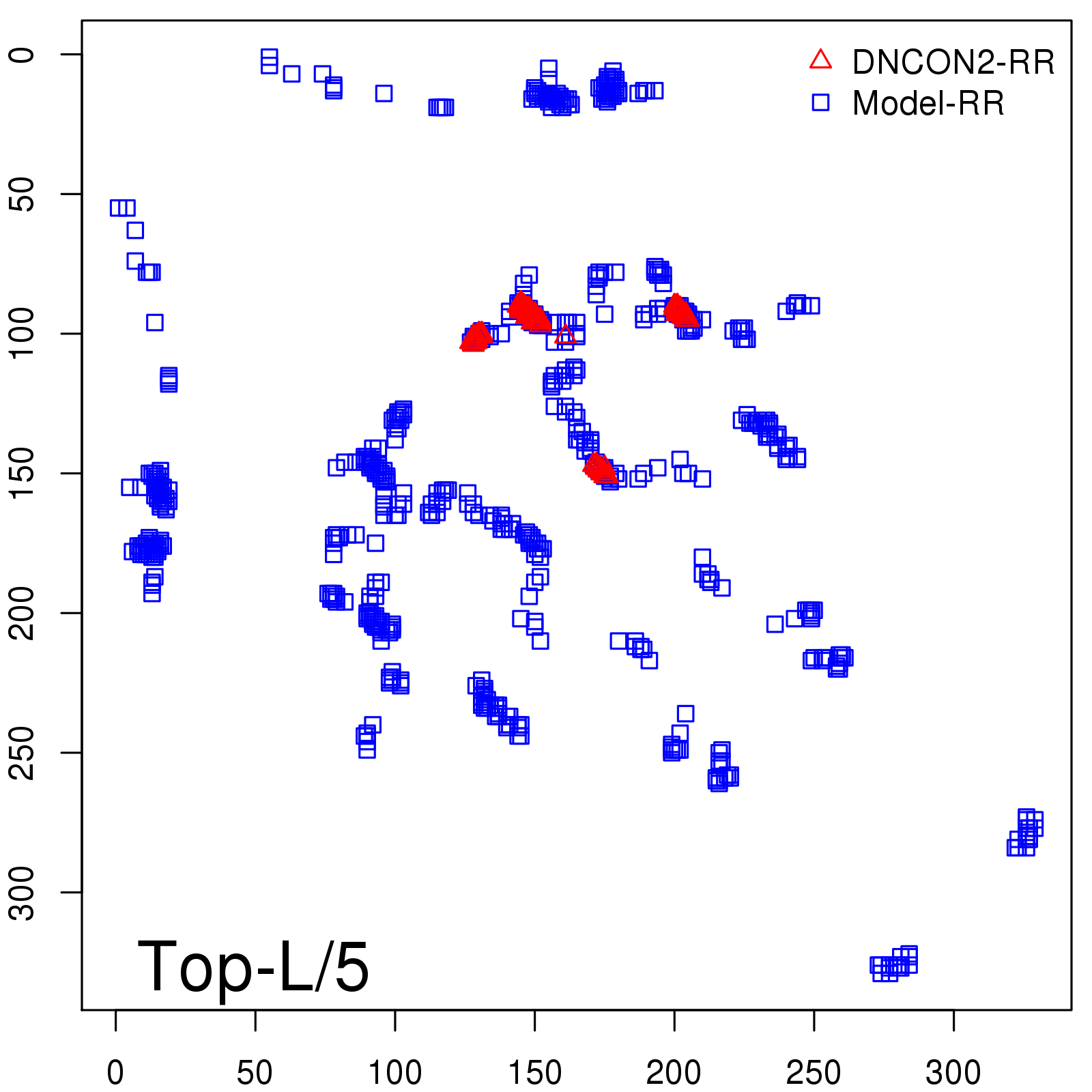
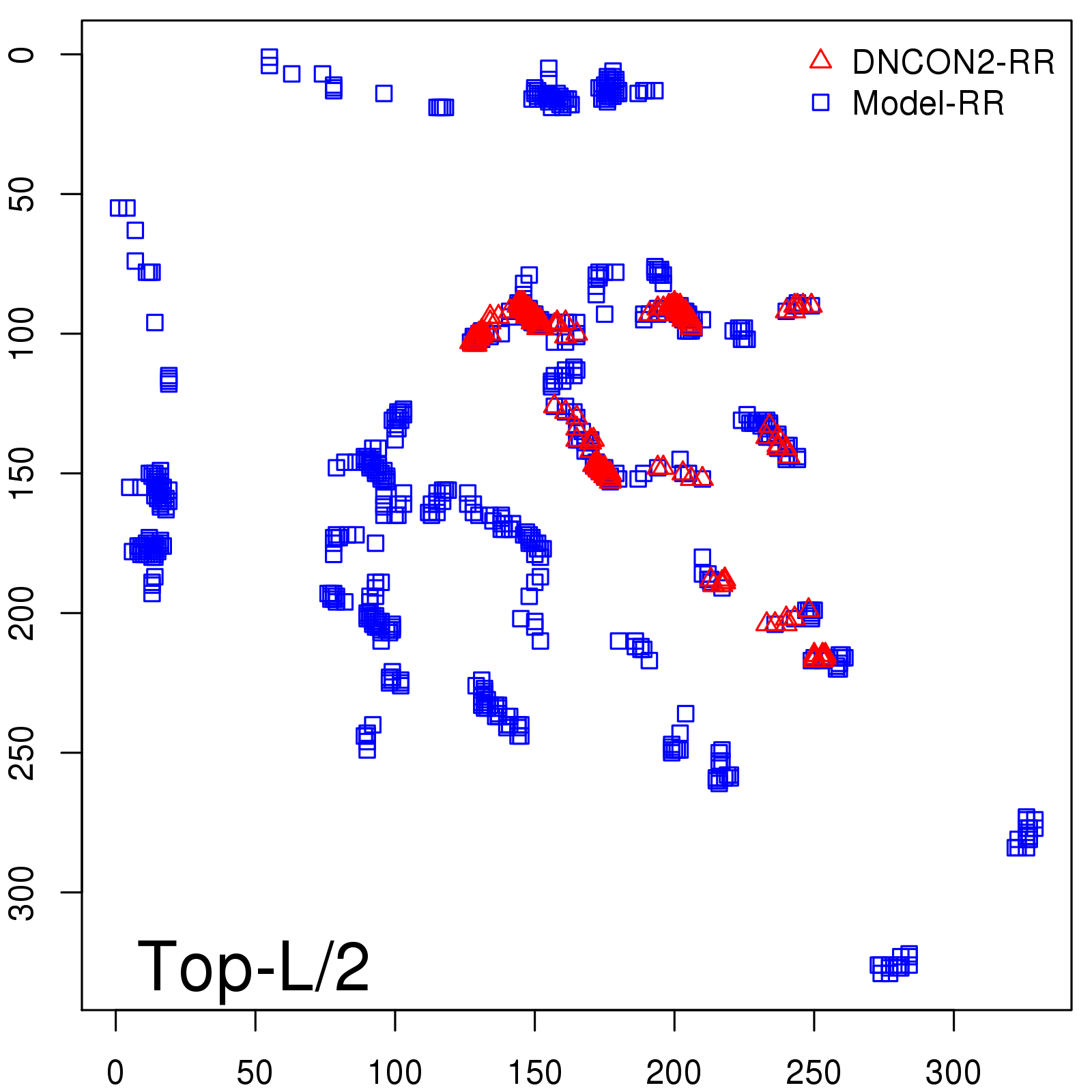
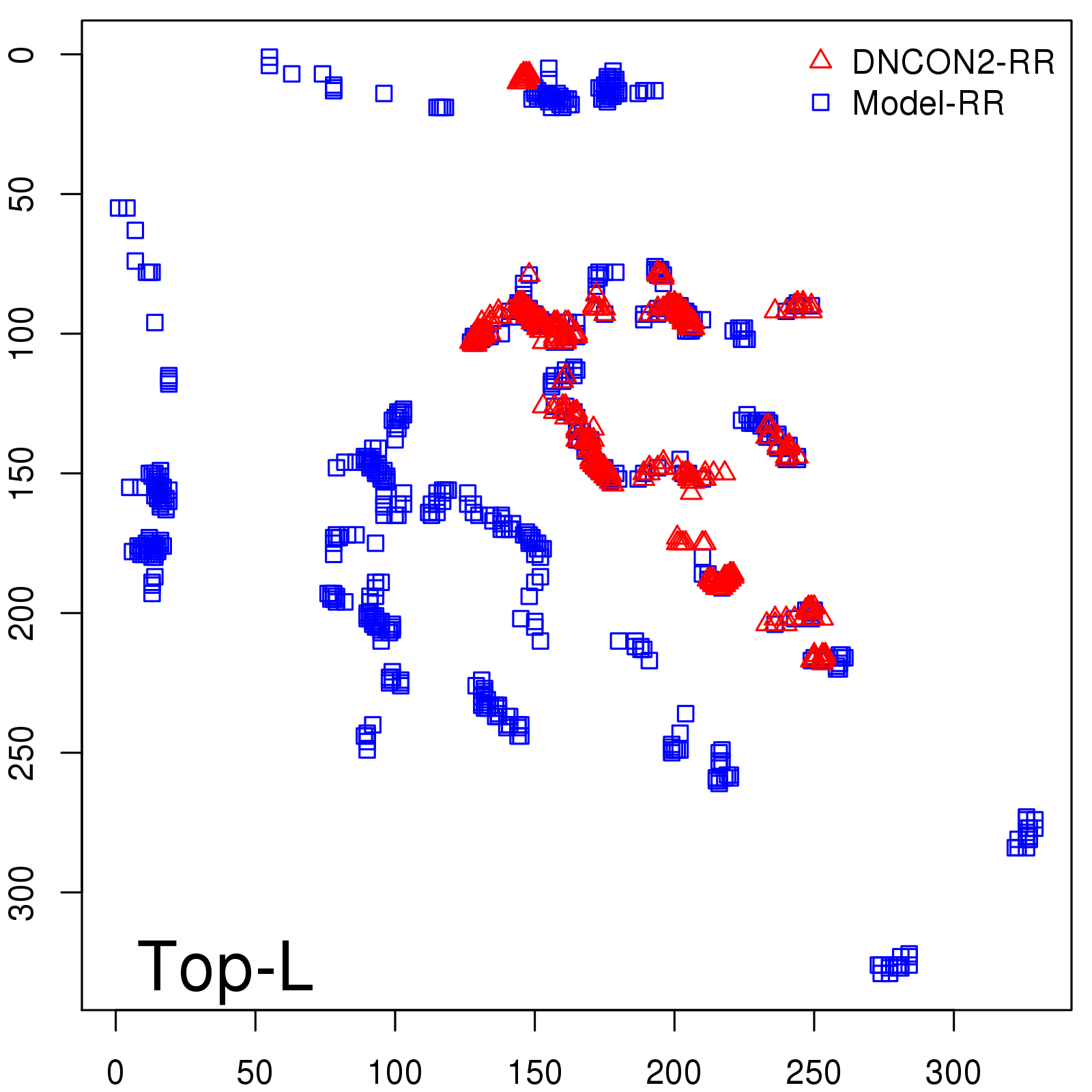
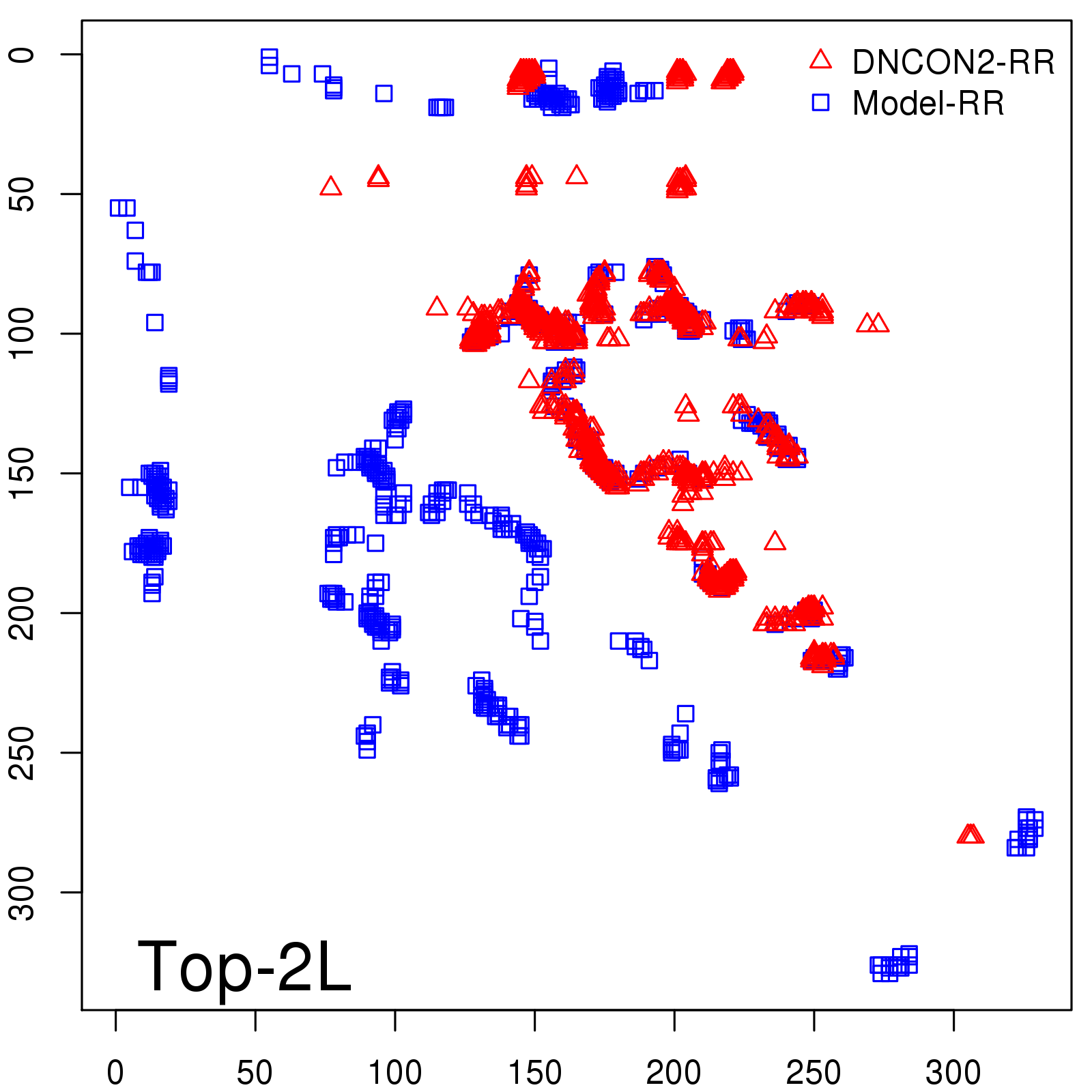
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 95.45 |
| TopL/2 | 74.55 |
| TopL | 48.94 |
| Top2L | 29.94 |
| Alignment | Number |
| N | 7193 |
| Neff | 280 |
Predicted Top 2 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 95.45 |
| TopL/2 | 73.94 |
| TopL | 48.94 |
| Top2L | 29.48 |
| Alignment | Number |
| N | 7193 |
| Neff | 280 |
Predicted Top 3 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 95.45 |
| TopL/2 | 75.76 |
| TopL | 49.85 |
| Top2L | 30.09 |
| Alignment | Number |
| N | 7193 |
| Neff | 280 |
Predicted Top 4 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 95.45 |
| TopL/2 | 74.55 |
| TopL | 48.94 |
| Top2L | 29.94 |
| Alignment | Number |
| N | 7193 |
| Neff | 280 |
Predicted Top 5 Tertiary structure
|
|
Predicted Contact Accuracy
|
| Long-Range | Precision |
| TopL/5 | 92.42 |
| TopL/2 | 72.73 |
| TopL | 48.02 |
| Top2L | 28.72 |
| Alignment | Number |
| N | 7193 |
| Neff | 280 |